1. Introduction
The BrainSuite BIDS App is a containerized software platform that provides a portable, streamlined mechanism for performing BrainSuite (https://brainsuite.org) analysis workflows to process and analyze anatomical, diffusion, and functional MRI data. With the BrainSuite BIDS App, you will be able to perform end-to-end neuroimaging analyses using its subject-level processing, quality control system, and hypothesis testing functionalities.
The BrainSuite BIDS app container includes complete R and Python environments, as well as all necessary packages and dependencies (e.g., the MATLAB Runtime library, FSL, and AFNI) to perform the participant-level processing and group-level analyses. This eliminates the need for you to install and configure environments or dependencies required to run BrainSuite tools, and does not interfere with the local environment on your host machine. The only installation you need to run to use the BrainSuite BIDS App is a container technology (Docker or Singularity).
Additionally, the BrainSuite BIDS App is designed to improve reproducibility in your analyses. The version-controlled BrainSuite BIDS app image ensures a consistent set of packages and prevents differences in outputs that are caused by varying installations of software versions. Furthermore, each time the software is run, subject-level processing parameters, software settings, and statistical model parameters are archived and saved in the output directories.
The current release of the BrainSuite BIDS App is based on version 23a of BrainSuite. The BrainSuite BIDS App implements three major BrainSuite pipelines for participant-level analysis, as well as corresponding group-level analysis functionality. For a detailed description of the BrainSuite BIDS App, please see our bioRxiv preprint (Kim et al., 2023).
1.1. Development Team
The BrainSuite BIDS App lead developer is Yeun Kim in the Shattuck Lab at the Ahmanson-Lovelace Brain Mapping Center at UCLA. The project is supervised by David Shattuck, with contributions by Shantanu Joshi, Anand Joshi, and Soyoung Choi.
The BrainSuite BIDS App is based on BrainSuite, an open-source software project that is produced and distributed as a collaborative effort led by David Shattuck at UCLA and Richard Leahy at the Biomedical Imaging Group at the University of Southern California. Major contributors to the BrainSuite project include Chitresh Bhushan, Soyoung Choi, Hanna Damasio, Justin P. Haldar, Anand A. Joshi, Shantanu H. Joshi, Yeun Kim, Divya Varadarajan, and Jessica L. Wisnowski.
1.2. Support
For questions or support, please visit the BrainSuite BIDS App forum or email us at support@brainsuite.org. You can also submit issues to our issue tracker on Github.
1.3. BrainSuite BIDS App Architecture
The BrainSuite BIDS App follows the BIDS App methodology and includes a participant-level workflow and a group-level analysis workflow. These are integrated as illustrated in Fig 1.
Participant-level processing requires BIDS-compatible input data. It can be run in parallel by launching multiple separate BrainSuite BIDS App containers or be run serially using a single container. While processing is running, the Dashboard can be launched as a separate BrainSuite BIDS App instance, which will generate a browser-based visualization QC page showing the progress of the processing in real-time. Once all the processing has completed, group analysis can be launched using another instance of the BrainSuite BIDS App.
Each workflow component can be configured using JSON files, which are described in the sections Modifying participant-level preprocessing parameters and Modifying group-level analysis parameters.
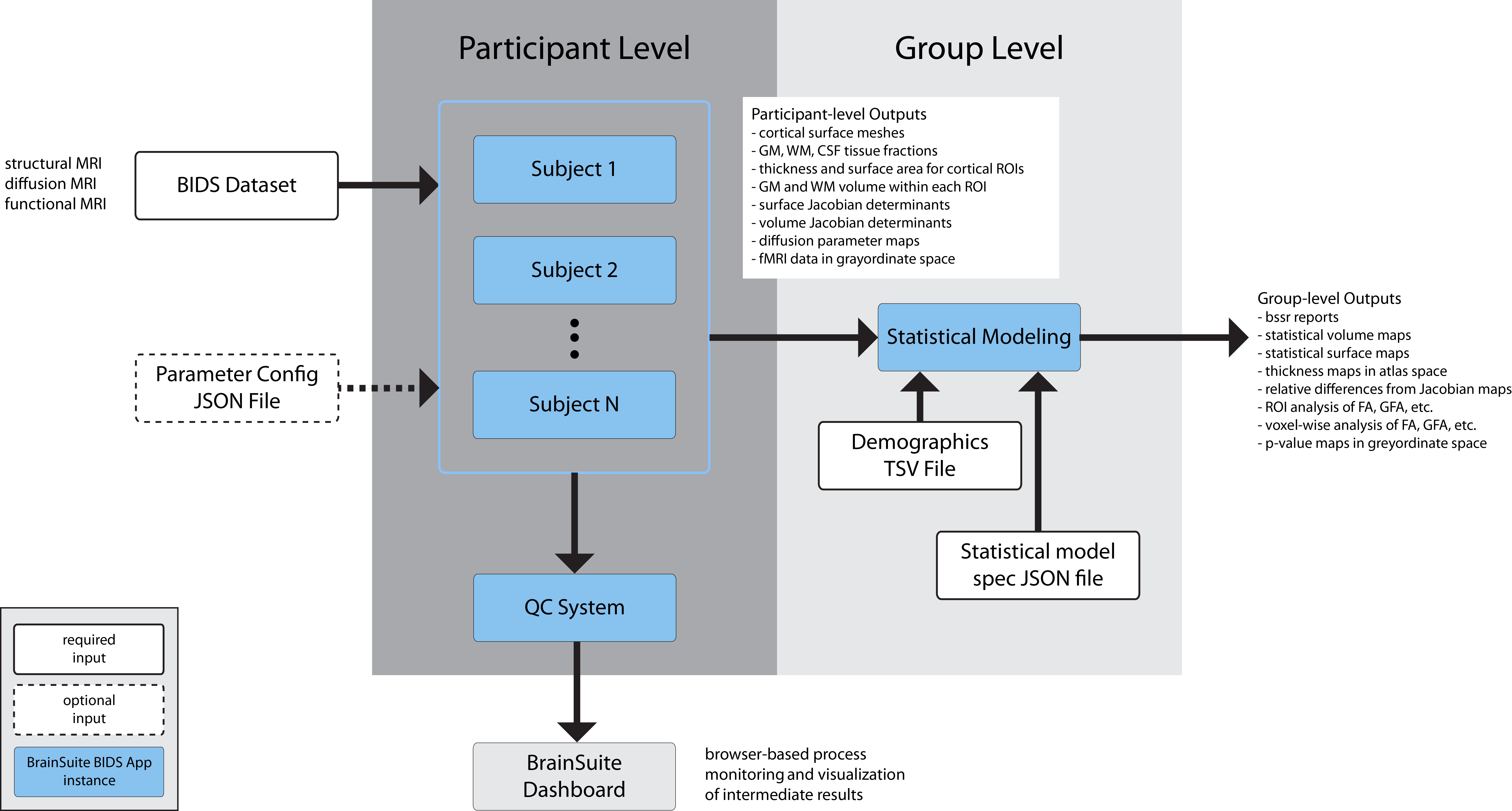
Fig. 1. BrainSuite BIDS App Architecture.
1.4. Participant-level Analysis
The participant-level workflow includes the Anatomical Pipeline, the Diffusion Pipeline, and the Functional Pipeline. These are each composed of several executable programs, most of which were developed as part of BrainSuite in the form of compiled C++ or MATLAB code. The BrainSuite BIDS App’s Diffusion Pipeline also uses FSL’s eddy, and its Functional Pipeline employs components from both AFNI and FSL.
The Anatomical Pipeline processes T1-weighted (T1w) MRI to compute vertex-wise cortical thickness measures and generate brain surfaces and volumes that are registered and labeled using surface-constrained volumetric restigration to align to a labeled reference anatomical atlas (CSE and SVReg).
The Diffusion Pipeline processes diffusion MRI (dMRI) and performs eddy current and motion correction (FSL’s eddy), coregistration of the dMRI data to the T1w data, distortion correction, diffusion model fitting, and DTI parametric map generation (BDP).
The Functional Pipeline processes resting-state and task-based functional MRI (fMRI) data and performs coregistration of the fMRI data to the T1w data, motion correction, outlier detection, and transformation of the data to the anatomical atlas space and to the grayordinate space (BFP).
For more details on the pipelines, please refer to the BrainSuite Pipelines page or our bioRxiv preprint (Kim et al., 2023).
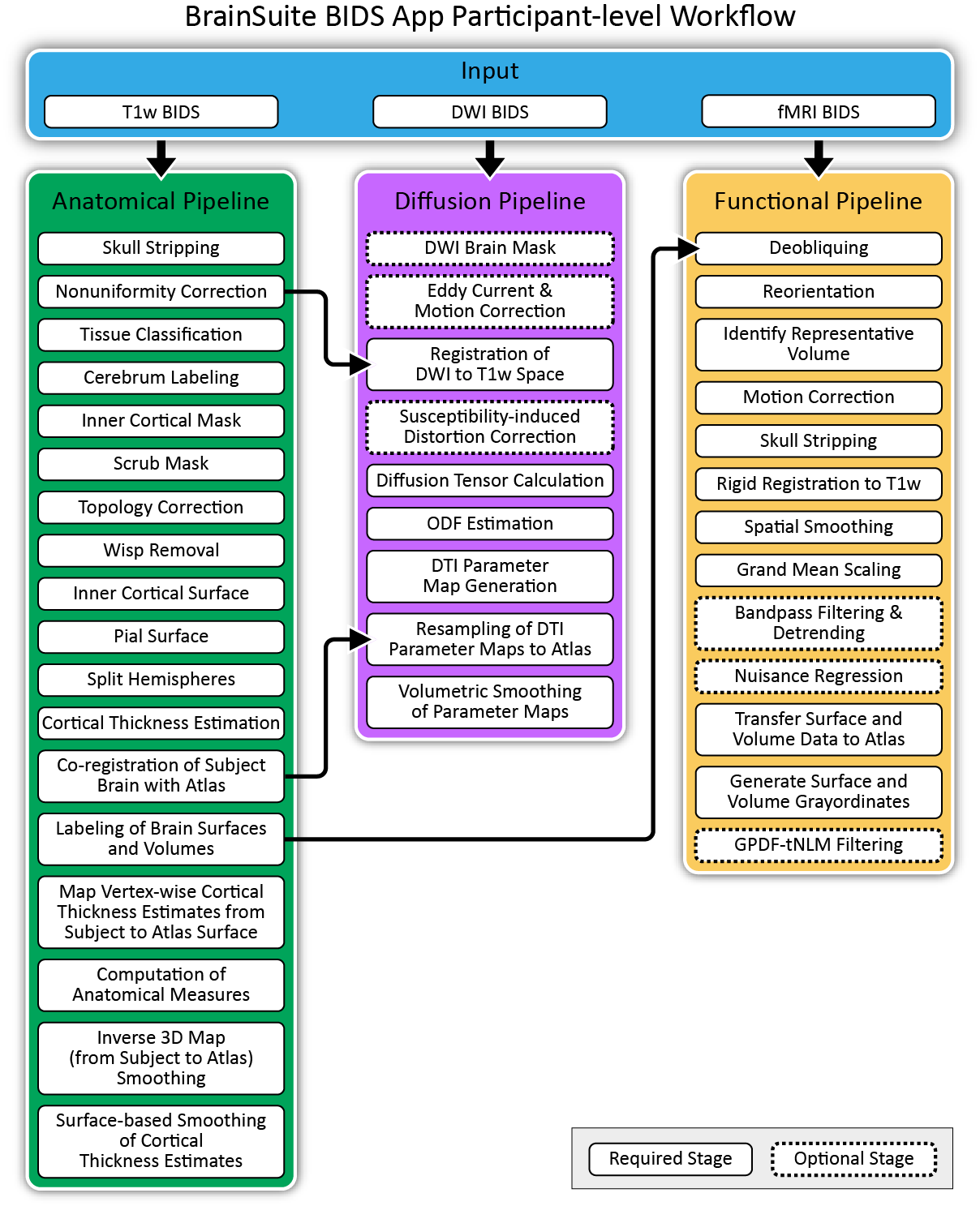
Fig. 2. Participant-level Workflows.
1.5. BrainSuite Dashboard
The intermediate outputs of these pipelines can be visualized using the BrainSuite Dashboard, a browser-based tool that lets you monitor your data processing as it happens. BrainSuite Dashboard provides snapshot images of several stages of the pipelines, which can help you detect processing errors or other issues. For more details on how to run the Dashboard, please see the BrainSuite Dashboard page. An example of the BrainSuite Dashboard is available at https://brainsuite.github.io/DashboardDemo/.
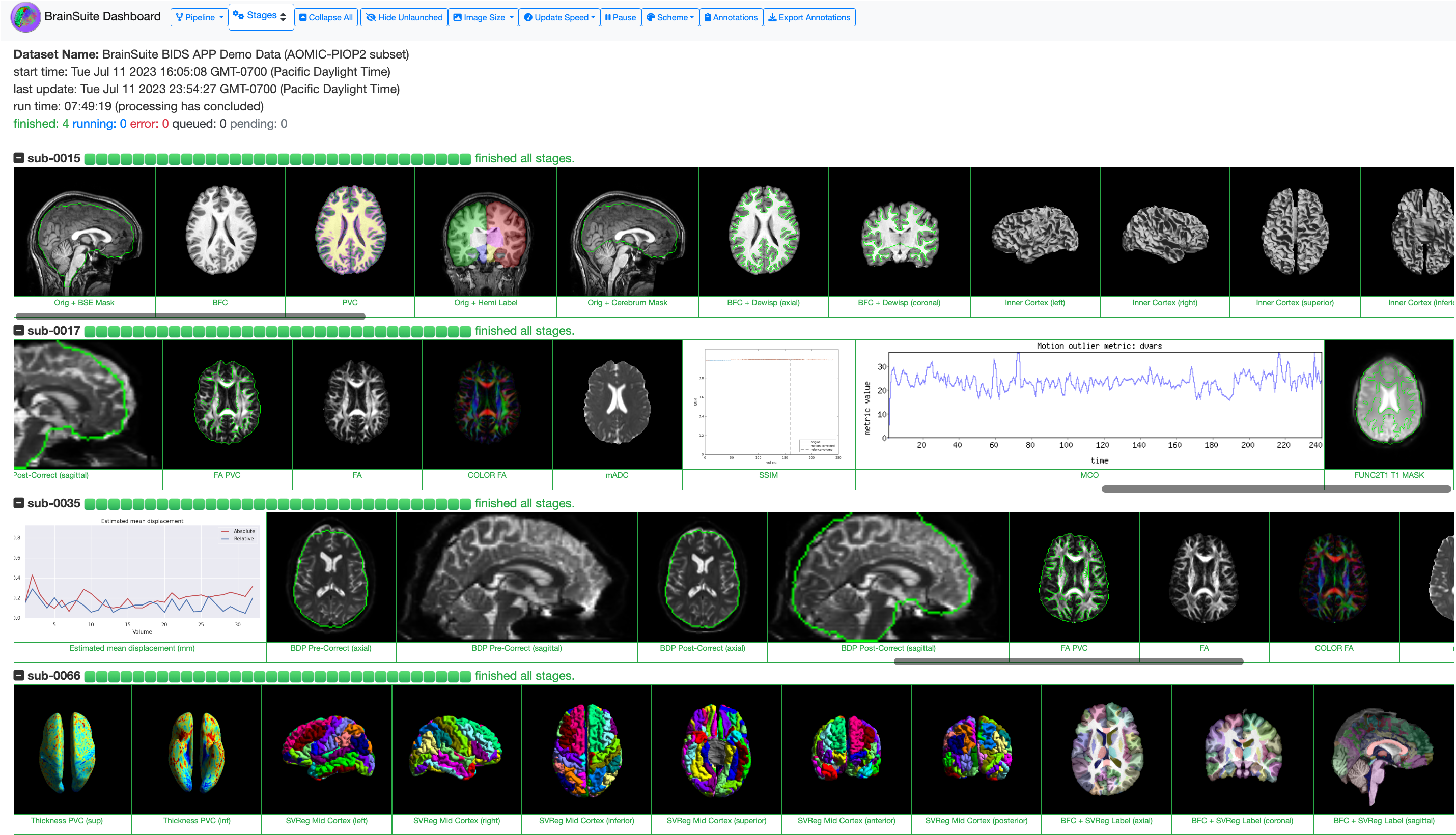
Fig. 3. The BrainSuite Dashboard.
1.6. Group-level Analysis
The group-level workflows use the BrainSuite Statistics Toolbox in R (bstr) to analyze the structural MRI outputs and BrainSync and Python-based tools to analyze the functional MRI outputs. Table 1 shows the variety of analyses that are available in the BrainSuite BIDS App: For more details regarding group-level stage workflows, please see Group-level Analysis Workflows page.
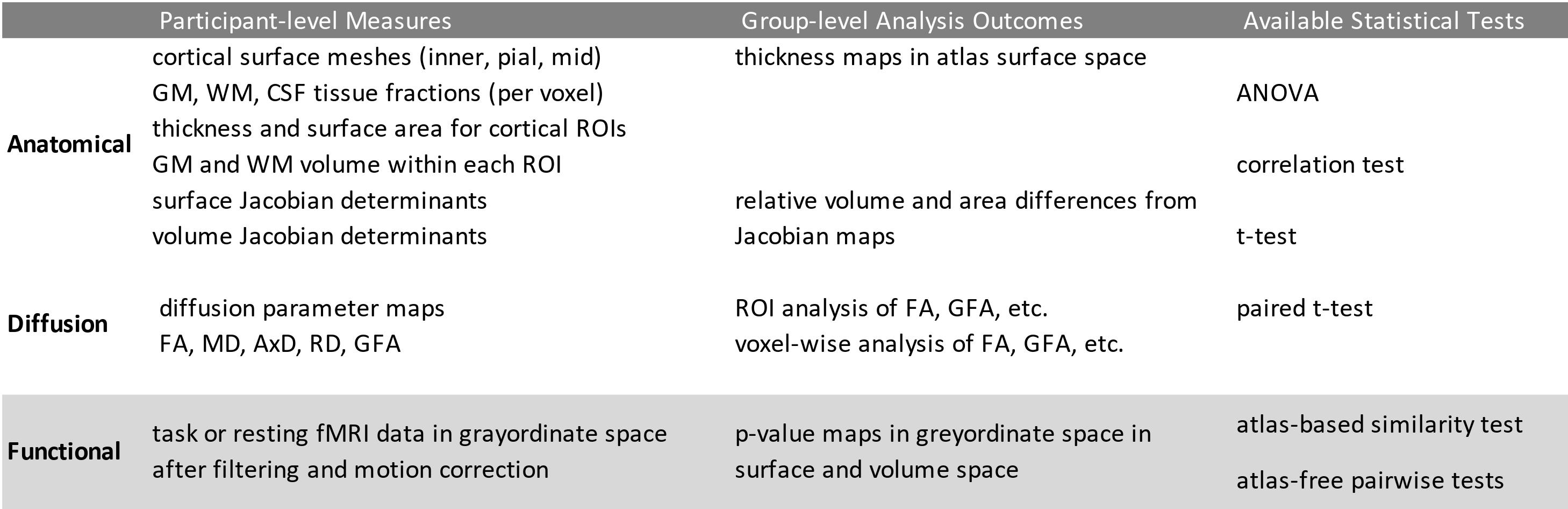
Table 1. Available analyses in BrainSuite BIDS App.
1.6.1. Group-level Analyis of Anatomical and Diffusion Data
bstr supports the following methods for performing group-level statistical analysis:
tensor based morphometry (TBM) analysis of voxel-wise magnitudes of the 3D deformation fields of MRI images registered to the atlas
cortical surface based analysis (SBA) of the vertex-wise thickness in the atlas space
diffusion parameter map (e.g., fractional anisotropy, mean diffusivity, radial diffusivity) based analysis (DBA)
region of interest (ROI)-based analysis of average gray matter thickness, surface area, and gray matter volume within cortical ROIs
Automated reports are generated by bstr (within the BrainSuite BIDS App) for TBM, SBA, and ROI-based analysis (see Fig. 4).
1.6.2. Group-level Analysis of Functional Data
Group-level statistical analysis of fMRI data (functional connectivity) is performed using BrainSync, a tool that temporally aligns spatially registered fMRI datasets for direct timeseries comparisons between subjects. After BrainSync, Python tools and scripts are called to perform statistical tests. Methods available for fMRI analyses are:
atlas-based linear modeling using a reference dataset created from multiple input datasets
atlas-free pairwise testing of all pairs of subjects is performed and used as test statistics for regression or group difference studies
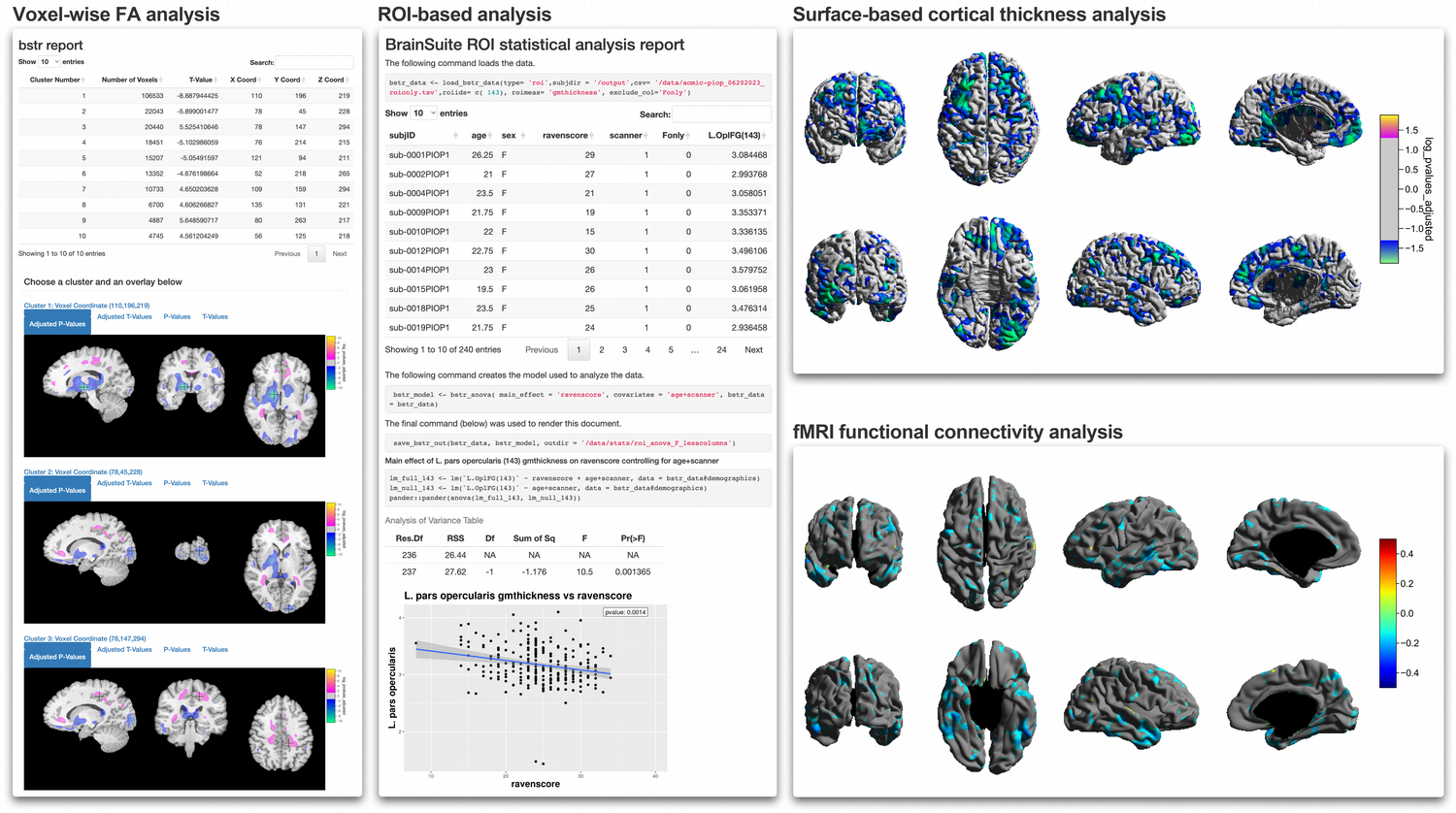
Fig. 4. Group Analyis
1.7. Licenses
The primary BrainSuite BIDS App source code is licensed under the GNU Public License v2.0 only (GPL-2.0-only).
The BrainSuite BIDS App makes use of several freely available software packages. Details on the licenses for each of these are provide in the files within the LICENSES directory of the BrainSuite BIDS App Github repository.
1.8. Acknowledgements
This project is supported by National Institutes of Health grants R01-NS074980, R01-NS121761, and R01-EB026299.
1.9. References
Related BrainSuite Papers:
[Kim2023a] The BrainSuite BIDS App.
[Kim2023b] BrainSuite BIDS App bioRxiv preprint.
[Joshi2023bstr] MRI Volumetric and Surface Data Analysis with the BrainSuite Statistics in R (bstr) Toolbox.
[Kim2018] BrainSuite BIDS App: a Containerized Version of the BrainSuite Processing Pipelines.
[Shattuck2001a] Skull stripping, bias field correction, tissue classification.
[Shattuck2001b] Topology correction.
[Shattuck2002] Skull stripping, bias field correction, tissue classification, topology correction, surface generation, GUI.
[Bhushan2015] INVERSION: Co-registration and Distortion Correction of Diffusion and Anatomical Images.
[Pantazis2010] Comparison of landmark-based and automatic methods for cortical surface registration.
[Joshi2007] Surface-Constrained Volumetric Brain Registration.
[Joshi2020bssr] The BrainSuite Statistics in R (bssr) Toolbox.
[Joshi2018bfp] BrainSuite fMRI Pipeline.
[Joshi2018ale] Cortical Thickness using the Anisotropic Diffusion.
[Joshi2022] USC Brain Atlas.