BrainSuite23a Released
We are pleased to announce the release of BrainSuite23a (release date: 23 July 2023). The latest release of BrainSuite includes contributions from David W. Shattuck, Chitresh Bhushan, Soyoung Choi, Hanna Damasio, Justin P. Haldar, Anand A. Joshi, Shantanu H. Joshi, Yeun Kim, Kayla A. Schroeder, Divya Varadarajan, Jessica L.
Wisnowski, and Richard M. Leahy.
One major change is that BrainSuite23a now uses the 2023a Matlab Runtime. This produces some minor differences in outputs, and we recommend not mixing results computed with BrainSuite 2023a with previous versions.
Please note that you may receive security warnings when installing or running the BrainSuite programs. This is likely to happen with newer versions of Mac OS. The BrainSuite dmg file is codesigned, and we are working to have it notarized by Apple to address this.
BrainSuite BIDS App v23a
- In conjunction with the release of BrainSuite23a, we have released an updated version of the BrainSuite BIDS App
- The BrainSuite BIDS App is a containerized environment that enables you to run the BrainSuite environment using Docker or Singularity. All system requirements are included in the Docker, facilitating easy installation.
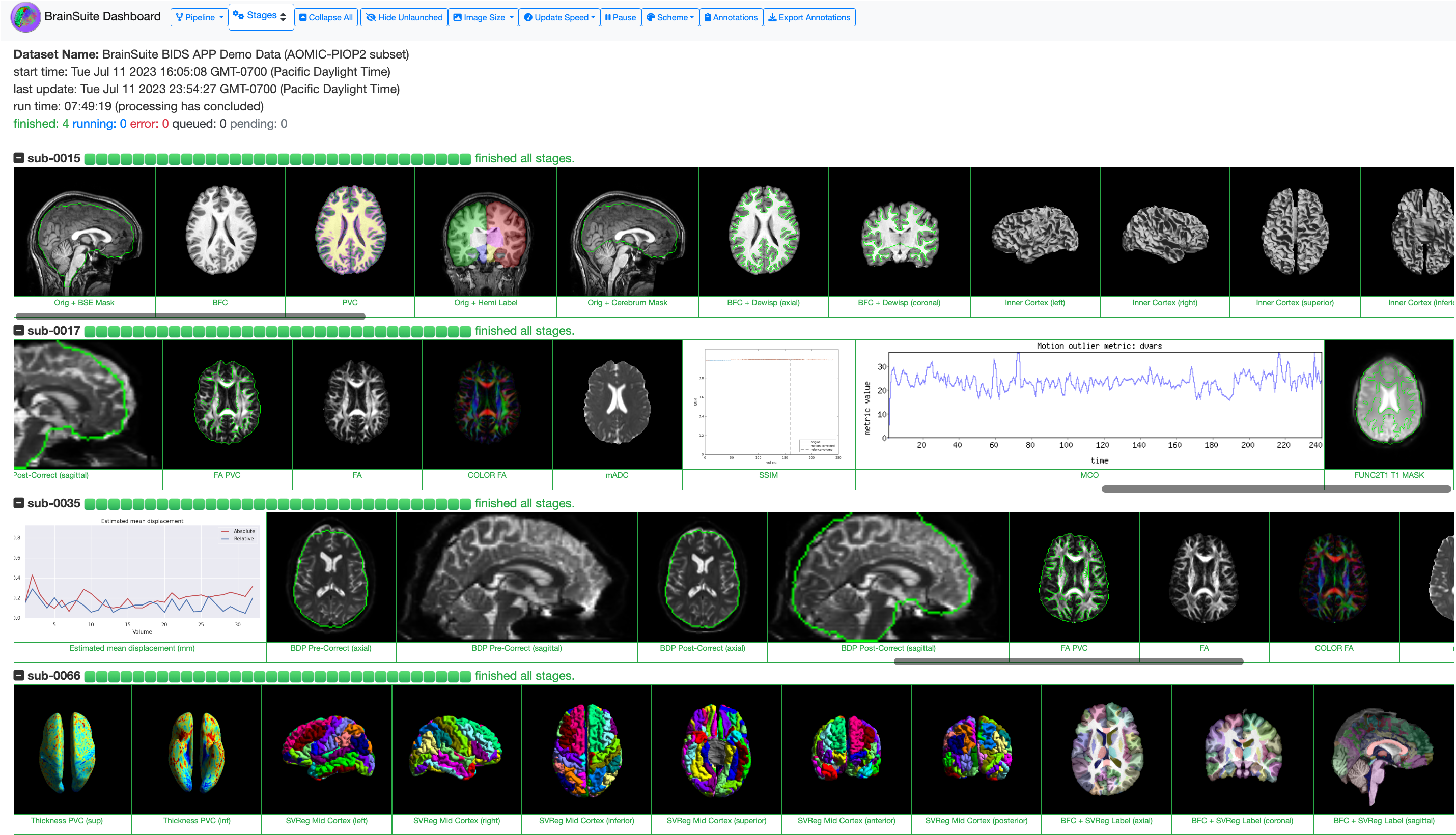
- The BrainSuite BIDS App includes BrainSuite Dashboard, a web-based system for monitoring the status of your study data as they are processed. This enables you to see errors and assess the quality of the derived data as they are created.
- The BrainSuite BIDS App does not include the BrainSuite GUI, which we recommend installing on your host machine if you are using Docker or Singularity for processing your data.
BrainSuite Statistics Toolbox in R (bstr)
- bssr had been renamed to bstr
- Updates to support BIDS compatibility
- Support excluding subjects from csv using the parameter exclude_col
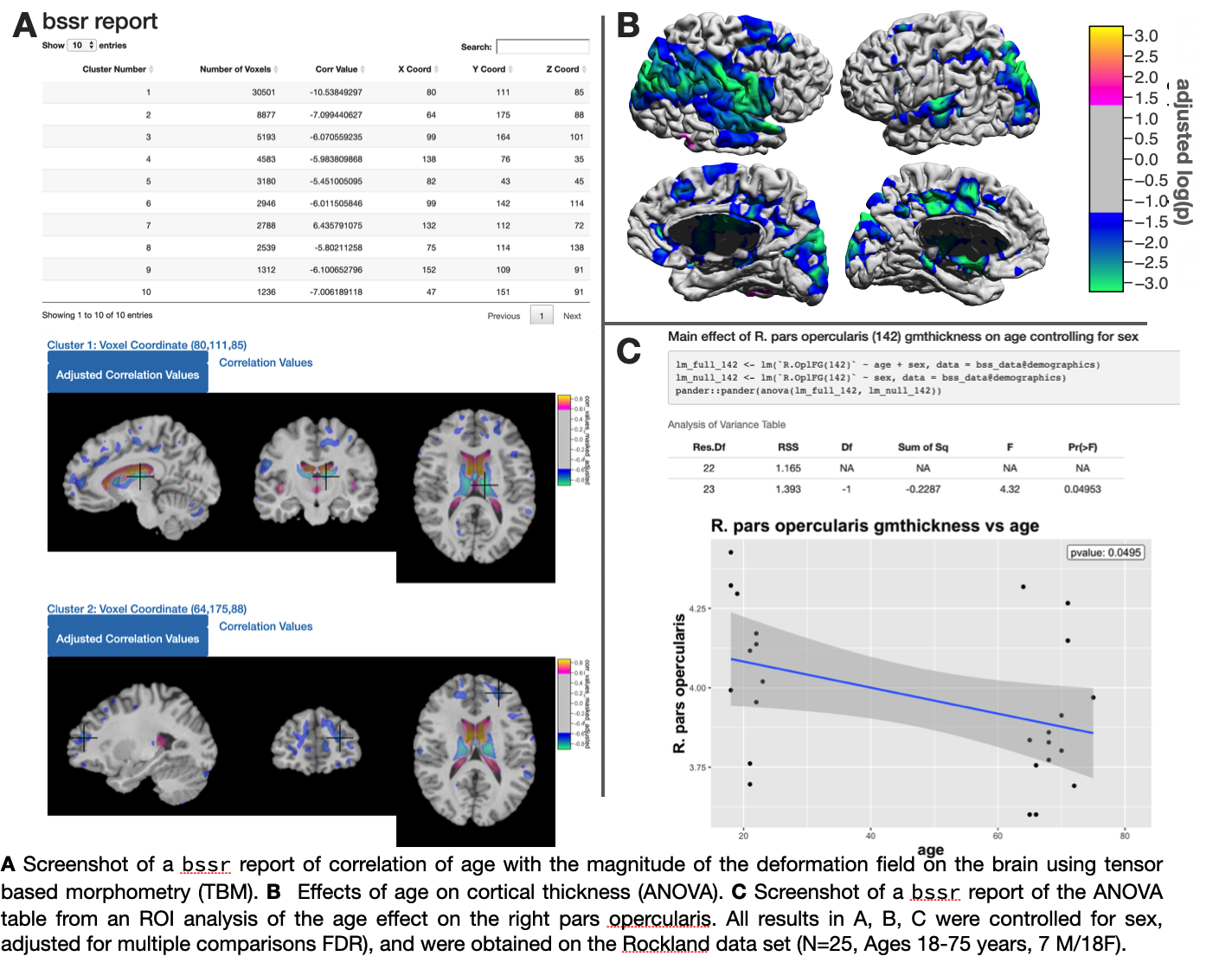
The BrainSuite Statistics Toolbox in R (bstr) contains new features for computing and plotting results.
BrainSuite Anatomical Pipeline
(new since 21a) The most major change to the software is a modification to the BrainSuite T1-weighted MRI processing workflow, which we have now termed the BrainSuite Anatomical Pipeline. The BrainSuite Anatomical Pipeline performs cortical surface extraction, cortical thickness estimation, surface-constrained volumetric registration, mapping of subject data to atlas space, and computation of subject-level statistics. We have separated the previous svreg program into multiple modules to improve the workflow.
We have developed new scripts to simplify batch processing of data. In the BrainSuite23a/bin folder, there is a script named brainsuite_anatomical_pipeline.sh (Mac/Linux) or brainsuite_anatomical_pipeline.cmd (Windows) which performs the full anatomical processing sequence. We have also created svreg_runall.{cmd,sh} which performs four steps using programs in the svreg bin folder: (cortical thickness estimation, surface-constrained volumetric registration, mapping of subject data to atlas space, and computation of subject-level statistics). The brainsuite_anatomical_pipeline script calls the cortical_surface_extraction and svreg_runall scripts.
BrainSuite Anatomical Pipeline Dialog
These changes to the workflow are also reflected in the BrainSuite Anatomical Pipeline Dialog, which is an update of the previous Cortical Surface Extraction dialog. The BAP Dialog also includes a new rewind feature. If you use the Automatically Save Results feature, then BrainSuite can rewind to a previous step by pressing on the rewind button (backwards circle arrow) next to the step name. This will rewind to the start of that step. This feature can also be used after closing and reopening BrainSuite.
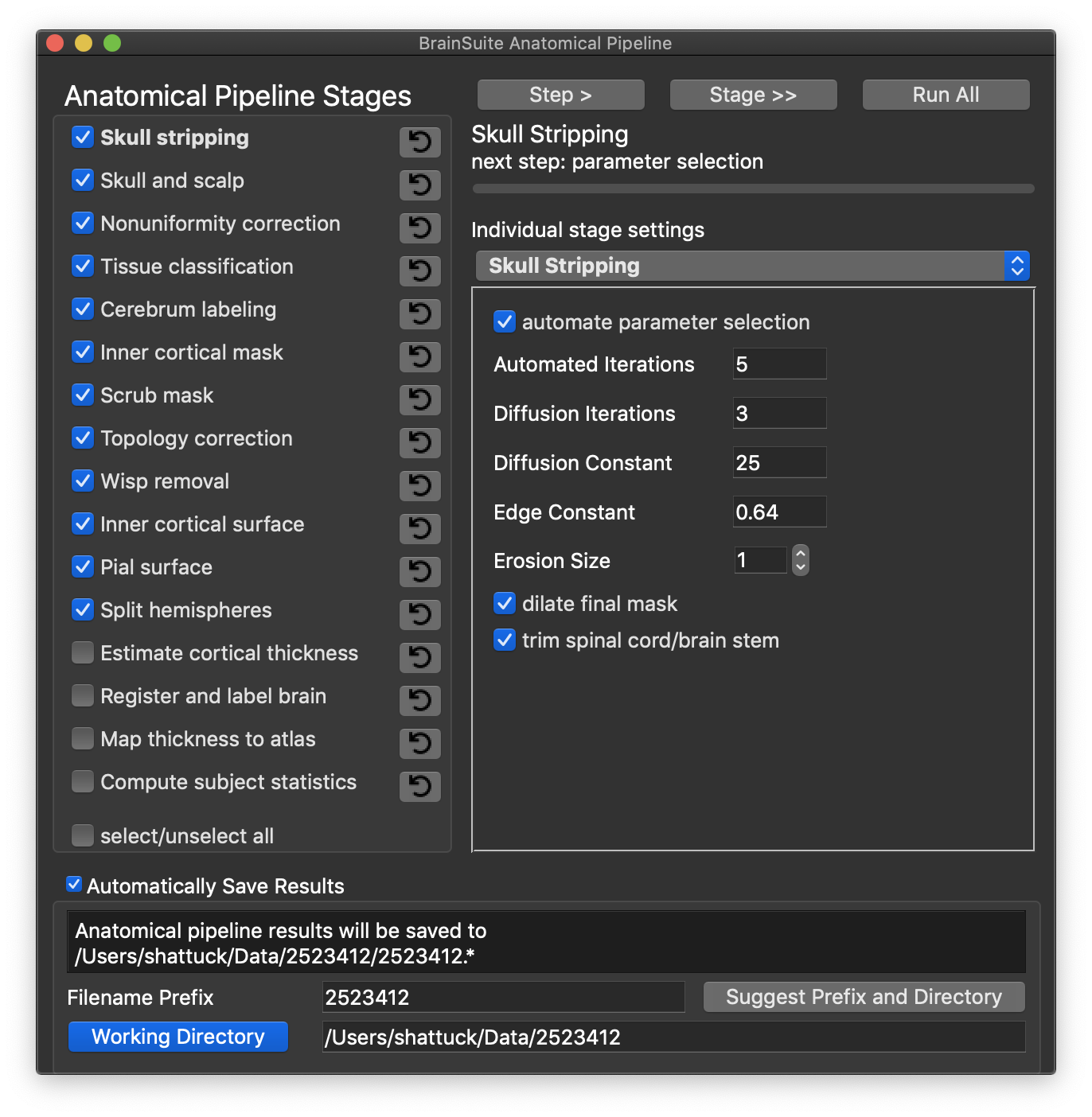
The BrainSuite Anatomical Pipeline dialog. This dialog, an updated version of the previous Cortical Extraction Sequence dialog, includes a new rewind feature. This enables the user to step back to previous steps in the toolchain. This can also be used to reload data after closing and reopening BrainSuite.
USCBrain Atlas
The USCBrain atlas is now included as part of the BrainSuite release. The USCBrain atlas is a new high-resolution single-subject atlas, constructed using both anatomical and functional information to guide the parcellation of the cortex. For details, please see the webpage for the atlas or our preprint describing the atlas, Joshi AA, Choi S, Chong M, Sonkar G, Gonzalez-Martinez J, Nair D, Wisnowski JL, Haldar JP, Shattuck DW, Damasio H, Leahy RM (2020) “A Hybrid High-Resolution Anatomical MRI Atlas with Sub-parcellation of Cortical Gyri using Resting fMRI” bioRxiv 2020.09.12.294322; doi: https://doi.org/10.1101/2020.09.12.294322
Surface-constrained Volumetric Registration (SVReg) Workflow
The SVReg workflow has changed. There are now four separate executables:
thicknessPVC– cortical thickness estimationsvreg– surface and volume registration and atlas-based labelingmapthickness– mapping thickness to atlas spacesvreg_thickness2atlas– computation of subject-level statistics (e.g., GM and WM volumes within anatomical ROIs)
These can be called from a single script (svreg_runall), which runs them in the above order.
Cortical Thickness Estimation
We have removed the use of linked distance as an estimate for cortical thickness. All BrainSuite processing for cortical thickness now uses thicknessPVC, an implementation of the Anisotropic Laplace Equation method described in Joshi et al., 2018. The ALE method provides much greater accuracy than linked distance, which was used previously to provide a fast visualization of thickness during cortical surface generation.
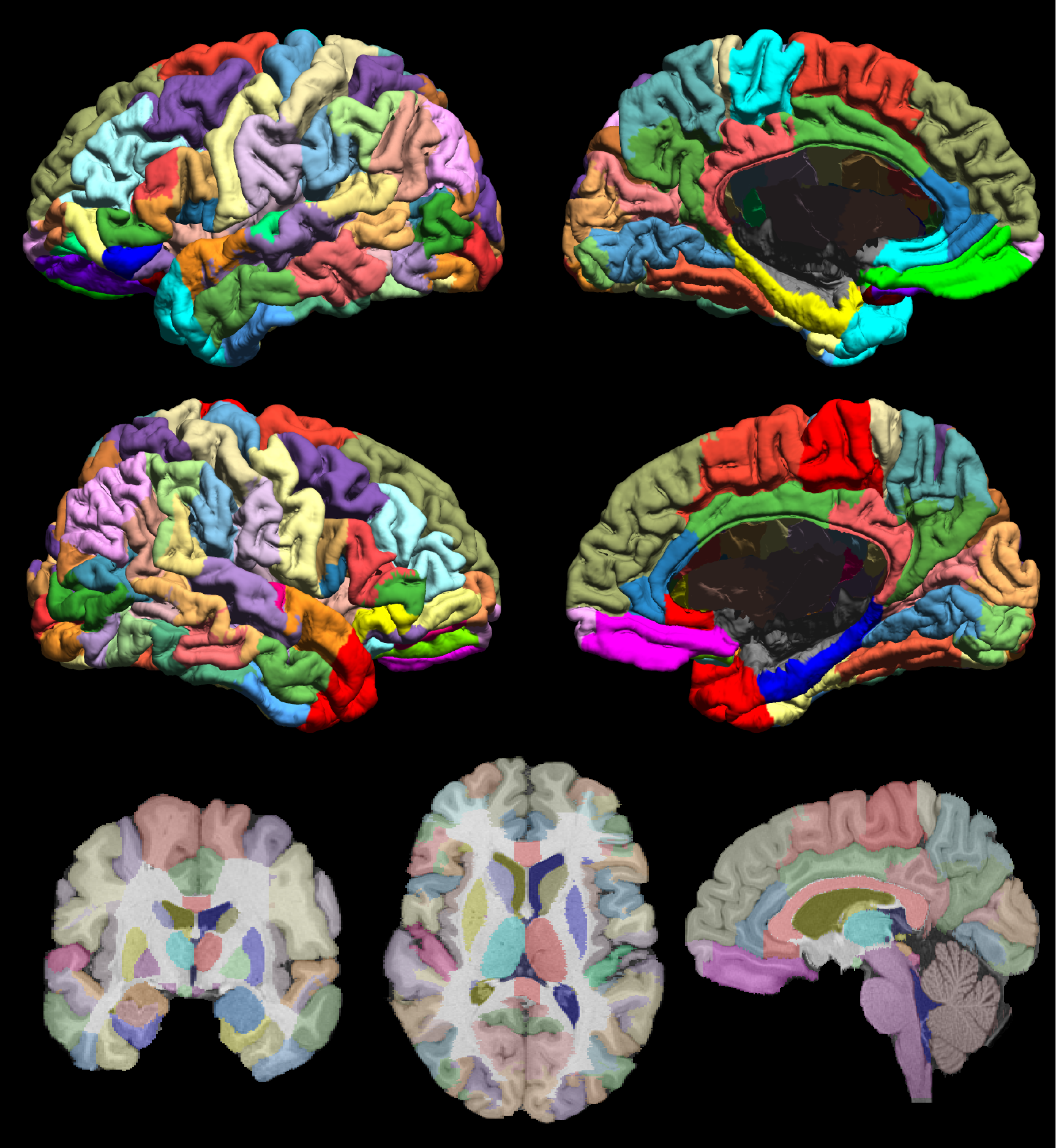
The USCBrain Atlas is now included in the BrainSuite21a release. This is a new high-resolution single-subject atlas, constructed using both anatomical and functional information to guide the parcellation of the cortex. The USCBrain Atlas consists of consistent volumetric and surface-based parcellations and labels. The volumetric component of the atlas contains a total of 159 regions of interest (130 cortical, 29 noncortical) defined with respect to the volumetric space. The surface-based component of the atlas has the same 130 sub-gyral (cortical) regions delineated and labeled in addition to the identification of 76 sulcal curves and delineation of gyral crowns and sulcal banks for each gyrus.