The BrainSuite BIDS App
Yeun Kim, Anand A. Joshi, Soyoung Choi, Shantanu H. Joshi, Chitresh Bhushan, Divya Varadarajan, Justin P. Haldar, Richard M. Leahy, and David W. Shattuck
To be presented at the 2023 Annual Meeting of the Organization for Human Brain Mapping (OHBM2023).
Introduction
We developed the BrainSuite BIDS App to provide containerized workflows for processing and analyzing anatomical, diffusion, and functional MRI data primarily using software we have developed for the BrainSuite project. By using the BIDS [1] and BIDS App standards [2], BrainSuite BIDS App can be rapidly deployed to provide a complete framework for performing end-to-end data analysis. We also introduce BrainSuite Dashboard, a browser-based quality control system that can be run concurrently with a set of BrainSuite BIDS App instances to facilitate real-time visual assessment of processing outputs.
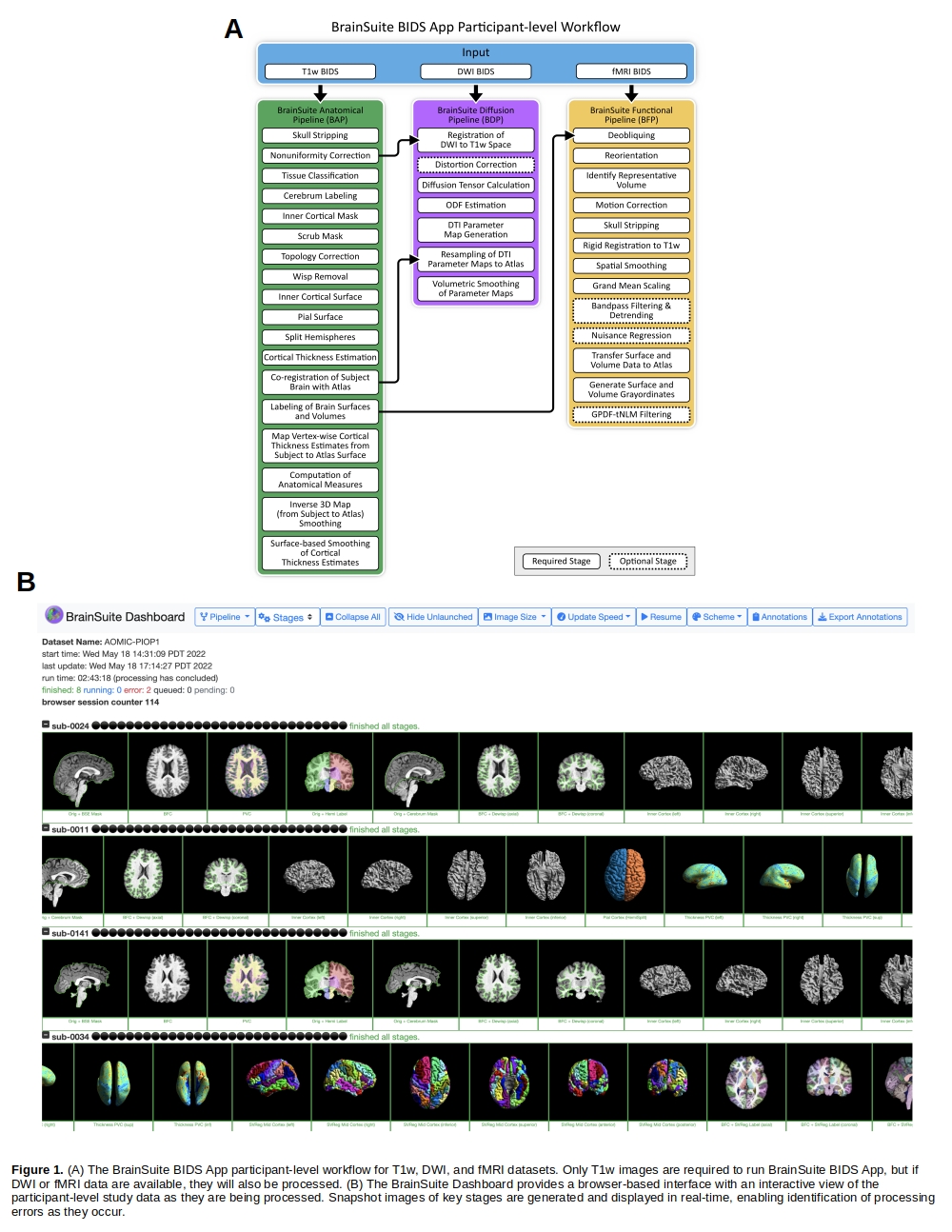
Methods
We implemented the BrainSuite BIDS App as a participant-level workflow comprising three pipelines (Fig. 1A) and a corresponding set of group-level analysis workflows. The BrainSuite Anatomical Pipeline (BAP) extracts cortical surface models from a T1-weighted (T1w) MRI [8], computes cortical thickness [3], and performs surface-constrained volumetric registration to align the T1w MRI to a labeled anatomical atlas [4]. The BrainSuite Diffusion Pipeline (BDP) processes diffusion MRI (dMRI) data by co-registering the dMRI data to the T1w scan, correcting for geometric image distortion, and fitting diffusion models [10]. The BrainSuite Functional Pipeline (BFP) processes functional MRI (fMRI) data by coregistering the fMRI data to the T1w data, then transforming the data to the anatomical atlas space and to the grayordinate space using tools from BrainSuite, FSL (fsl.fmrib.ox.ac.uk), and AFNI (afni.nimh.nih.gov). Group-level BAP and BDP outputs are analyzed using the BrainSuite Statistics in R (bssr) toolbox. BFP outputs can be analyzed using atlas-based or atlas-free statistical analyses using BrainSync [5], which synchronizes time-series data temporally. We also developed the browser-based BrainSuite Dashboard system, which can be run concurrently with a set of subject-level instances to provide rapid review of output data as they are generated (Fig. 1B).
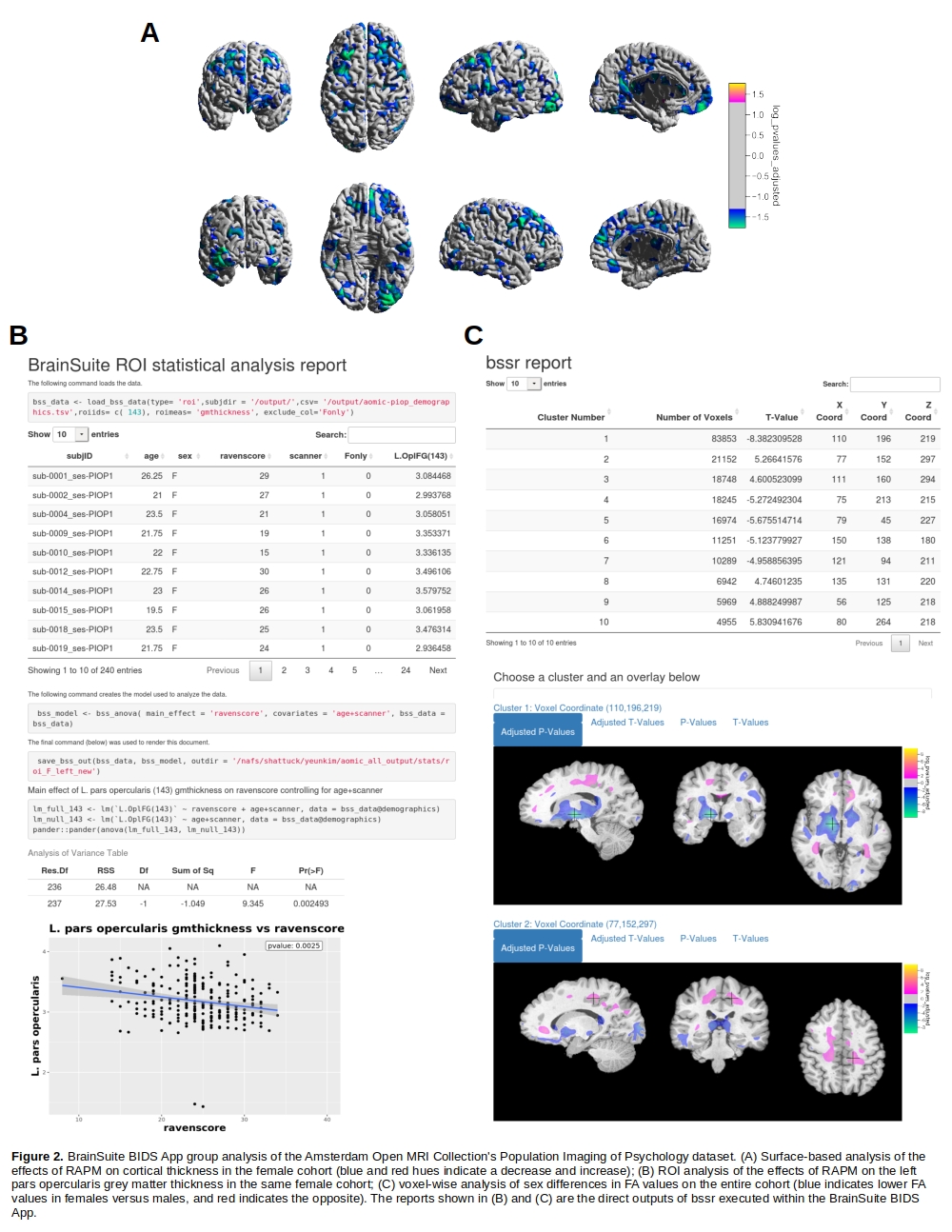
Results
As a demonstration of its utility, we applied the BrainSuite BIDS App to anatomical (T1w), diffusion, and functional MRI from the Amsterdam Open MRI Collection (AOMIC) Population Imaging of Psychology dataset [9]. After executing the participant-level workflows, we evaluated the outputs using BrainSuite Dashboard, adjusted software settings as needed, and reprocessed the data. We then performed five types of group-level analysis. Four of these examined effects of Raven’s Advanced Progressive Matrices (RAPM) scores, a proxy measure for intelligence, on: cortical thickness using surface-based analysis (SBA); volumetric change assessed using tensor-based morphometry (TBM); ROI analysis of left pars opercularis thickness (a region selected post hoc from the SBA result); and analysis of functional connectivity (FC) derived from resting-state fMRI. These analyses focused on the female cohort (N=240; age 22.07±1.74 years), in which we observed larger effect sizes compared to the male cohort. We also examined sex differences in fractional anisotropy (FA) values using a voxel-wise analysis (N=419; age 22.05±1.79 years; 240F/179M). After correcting for multiple comparisons, only SBA, ROI, and FA analyses produced statistically significant results. For SBA and ROI, we found decreased cortical thickness proportional to higher RAPM scores (Fig. 2A&B). In the FA analysis, we found higher FA values in the basal ganglia and lower FA values in the region below the postcentral gyri in males (Fig. 2C). Our findings were consistent with results from Schnack et al. [7] and Menzler et al. [6], who found similar cortical thickness and FA effects, respectively.
Conclusions
We developed the BrainSuite BIDS App and BrainSuite Dashboard and demonstrated their utility on the AOMIC dataset. These tools provide a practical mechanism for rapidly deploying BrainSuite workflows to perform large-scale studies on data organized according to the BIDS standard. More information can be found at http://brainsuite.org.
References
[1] Gorgolewski, K.J., et. al. (2016). The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Scientific Data, 3, 160044.
[2] Gorgolewski, K. J., et al. (2017). BIDS Apps: Improving ease of use, accessibility, and reproducibility of neuroimaging data analysis methods. PLoS Computational Biology, 13, e1005209.
[3] Joshi, A. A., et al. (2018). Using the anisotropic laplace equation to compute cortical thickness. In MICCAI 2018 (pp. 549–556).
[4] Joshi, A. A., et al. (2012). A method for automated cortical surface registration and labeling. In Biomedical Image Registration – 5th International Workshop, WBIR 2012, Nashville, TN, USA, July 7-8, 2012. Proceedings (pp. 180–189).
[5] Joshi, A. A., et al. (2021). A pairwise approach for fMRI group studies using the BrainSync transform. In: Medical Imaging 2021: Image Processing (p. 115960G). International Society for Optics and Photonics volume 11596.
[6] Menzler, K., et al. (2011). Men and women are different: diffusion tensor imaging reveals sexual dimorphism in the microstructure of the thalamus, corpus callosum and cingulum. NeuroImage, 54, 2557–2562.
[7] Schnack, H. G., et al. (2015). Changes in thickness and surface area of the human cortex and their relationship with intelligence. Cerebral Cortex, 25, 1608–1617.
[8] Shattuck, D. W., & Leahy, R. M. (2002). BrainSuite: An automated cortical surface identification tool. Medical Image Analysis, 8, 129–142
[9] Snoek, L., et al. (2021). The Amsterdam Open MRI Collection, a set of multimodal MRI datasets for individual difference analyses. Scientific Data, 8.
[10] Varadarajan, D., et al. (2020). Brainsuite Diffusion Pipeline (BDP): Processing tools for diffusion-MRI. 25th Annual Meeting of the Organization for Human Brain Mapping. Virtual.