Pial Surface Generation
 This module takes as input the inner cortex mask and the surface defined by its boundary. It also uses a set of tissue fraction values that define how much GM, WM and CSF are contained within each voxel of the brain image. Using an iterative process, each vertex of the WM surface is moved under the influence of several forces. The first pushes the vertex outward along the surface normal while the other forces try to maintain the smoothness of the initial surface. The movement of each vertex stops when it moves into a location with a significant CSF component or when its motion would cause the surface to self-intersect. The result is a one-to-one map between the points on the inner cortical surface model and the pial surface model, which can provide a direct estimate of cortical thickness.
This module takes as input the inner cortex mask and the surface defined by its boundary. It also uses a set of tissue fraction values that define how much GM, WM and CSF are contained within each voxel of the brain image. Using an iterative process, each vertex of the WM surface is moved under the influence of several forces. The first pushes the vertex outward along the surface normal while the other forces try to maintain the smoothness of the initial surface. The movement of each vertex stops when it moves into a location with a significant CSF component or when its motion would cause the surface to self-intersect. The result is a one-to-one map between the points on the inner cortical surface model and the pial surface model, which can provide a direct estimate of cortical thickness.
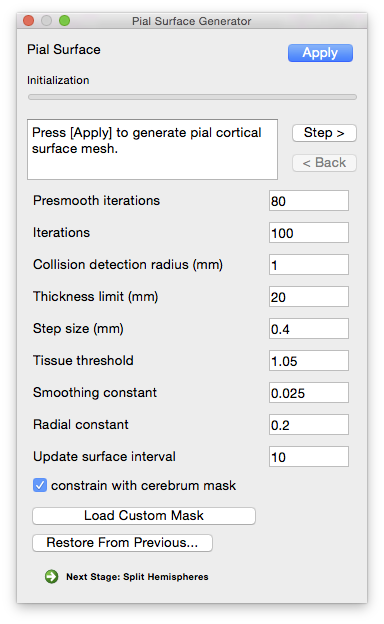
Parameters
- Presmooth iterations
- The number of iterations to run while smoothing the image before expansion. Increasing this number may help if the generated surface is too bumpy.
- Iterations
- The number of expansion iterations to run.
- Collision detection radius (mm)
- The size of the grid used for collision detection, in mm. Increasing this value increases the collision detection power of the tool, but at the expense of longer run times. Smaller values require more memory. Generally values less than 1 should be avoided to prevent the system from running out of memory.
- Thickness limit (mm)
- Determines the maximum distance (in mm) a vertex can travel during expansion. Setting this number to a high value, such as the default 20, effectively disables this functionality.
- Step size (mm)
- How far (in mm) a vertex moves each step.
- Tissue threshold (typical range: 1–1.5)
- The stopping point for motion based on the tissue classification map. A value of 1 corresponds to 100% CSF, while a value of 1.5 indicates 50% CSF / 50% grey matter. Decrease this value if the generated surface seems too conservative (i.e. it’s failing to expand to the outer surface of the brain). Increase this value if the generated surface is too expansive (if it expands beyond the apparent brain surface).
- Smoothing constant (range: 0–1)
- Determines the amount of smoothing ranging from 0 (no smoothing) to 1 (most smoothing)
- Radial constant
- Update surface interval
- Determines how often the surface view is updated during expansion (i.e. how many iterations before the view is updated to display the current progress). A value of 0 means that no intermediate results will be shown.
- Constrain with cerebrum mask
- If checked, keeps the pial surface from expanding outside of the volume enclosed by the cerebrum mask (e.g. expanding into the cerebellum). This stage uses the mask generated during the Cerebrum Labeling stage. To use a different mask (such as a manually edited one), click the “Load Custom Mask” button next to this item and select it.
Command-Line Usage
pialmesh: computes a pial surface model using an inner GM/WM mesh and a tissue fraction map.
usage: pialmesh -i input -o output -f fraction_file -m mask [optional settings]
example: pialmesh -i input_mesh.dfs -o output_mesh.dfs -f frac_file.nii.gz -m mask_file.nii.gz
Required Settings:
| Flags | Description |
|---|---|
-i <file> |
input mesh file |
-o <file> |
output mesh file |
-f <fraction file> |
floating point (32) tissue fraction image |
-m <mask file> |
restrict growth to mask file region |
Optional Settings:
| Flags | Description |
|---|---|
-g |
debug |
-v <level> |
verbosity [default: 0] |
-n <n> |
number of iterations [default: 10] |
-r <radius> |
search radius [default: 2] |
-s <step size> |
step size [default: 0.1] |
--max <thickness> |
maximum allowed tissue thickness [default: 10] |
-t <threshold> |
tissue threshold [default: 1.1] |
--interval <n> |
output interval [default: 0] |
--prefix <prefix> |
prefix for exporting surfaces if interval is set |
--smooth <smoothing constant> |
apply Laplacian smoothing [default: 0.1] |
--timer |
show timing |
--norm |
recompute normals at each iteration |
--nc <normal_constant> |
strength of normal smoother. [default: 0] |
--tc <tangential_constant> |
strength of tangential smoother. [default: 0] |
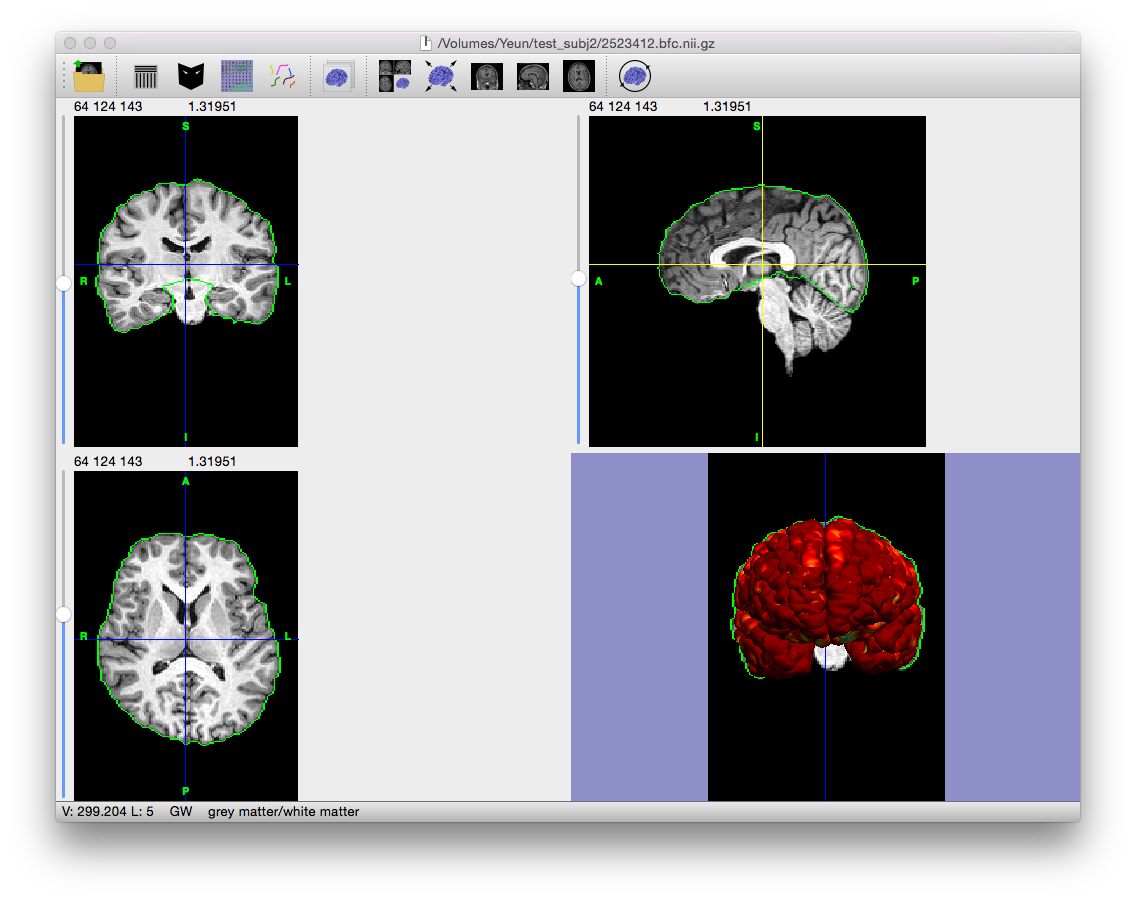
Example Result
Output File
If “save output of each stage automatically” is checked on the Cortical Surface Extraction dialog, the following file is generated (where filename_prefix is the filename of the MRI scan without the file extension, e.g. “testsubj” for the file “testsubj.nii”):
| Filename | Contents |
|---|---|
| filename_prefix.pial.cortex.dfs | Pial surface of the subject’s brain |
Restore from Previous Session
If BrainSuite was interrupted while performing this stage or to change the parameters for this stage and rerun after fully processing a scan, click “Restore From Previous…” on the bottom of this stage’s dialog box and load the original MRI scan. BrainSuite will automatically load all of the files generated in previous stages, allowing processing to restart from this intermediate stage.