bstr: BrainSuite Statistics Toolbox in R
Copyright (C) 2023 The Regents of the University of California
Created by Shantanu H. Joshi, Yeun Kim, Kayla A. Schroeder, David W. Shattuck
bstr is licensed under an GPLv2-only license (https://spdx.org/licenses/GPL-2.0-only.html).
The BrainSuite Statistics Toolbox in R (bstr) is a software package developed in R that performs statistical analysis of population-level neuroimaging data processed using BrainSuite [1]. Specifically, it provides statistical tools for conducting cortical thickness analysis, tensor based morphometry, and analysis of diffusion measures.
Installation
Prerequisites
Ensure BrainSuite is installed on your computer.
Ensure R is installed on your computer.
Install RStudio.
Install Rtools (Windows only).
Steps for installation
Open RStudio and enter the following two commands to install bstr version 0.4.1
install.packages('devtools')devtools::install_url('https://brainsuite.org/wp-content/uploads/2023/07/bstr_0.4.1.tar.gz')
Check your installation
Type library(bstr).
Then type get_brainsuite_install_path().
This should display the BrainSuite installation path.
Methods
A subject-level BrainSuite workow prior to conducting statistical analysis involves T1-weighted MRI image processing and registration steps, including cortical surface extraction [1] and alignment to a reference atlas using SVReg [2]. SVReg performs surface-constrained volumetric registration of triangular meshes and image intensities. bstr is then used to perform population level statistical analysis of various neuroimaging measures.
bstr supports the following analysis methods:
- tensor based morphometry (TBM) analysis of voxel-wise magnitudes of the 3D deformation fields of MRI images registered to the atlas
- cortical surface analysis of the vertex-wise thickness in the atlas space
- diffusion parameter maps analysis (e.g., fractional anisotropy, mean diffusivity, radial diffusivity). The statistical analysis is performed in a common coordinate space of an atlas by resampling the data from subject coordinates to a common atlas space using SVReg [2,3];
- region of interest (ROI)-based analysis of average gray matter thickness, surface area, and gray matter volume within cortical ROIs. It also offers tools for correcting for multiple comparisons using false discovery rate (FDR) or permutation testing methods.
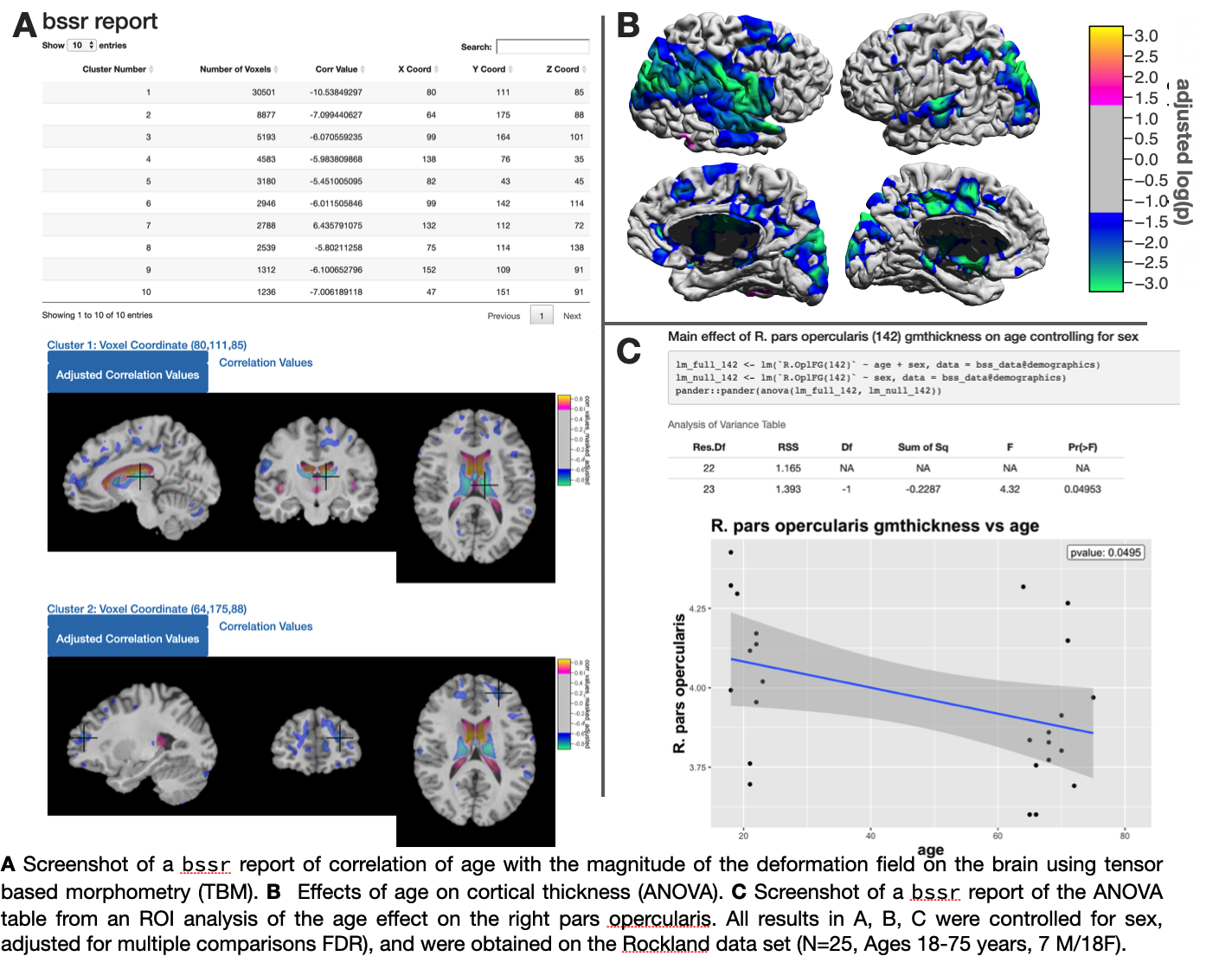
bstr is cross-platform and is available on macOS, Windows,and Linux based systems (all platforms with R support). bstr is distributed under an open source license (GPLv2-only). bstr supports functionality for automated report generation to visualize statistical results using R-shiny and R markdown. The volumetric analysis report contains the cluster table, visualizations of clusters on image slices, and shows both the unadjusted and the adjusted versions of p-values and t statistics, respectively. The ROI analysis report shows the demographic spreadsheet, automatic bar plots for ANOVA and regressions, and scatter plot for correlation analyses. bstr also exports an R markdown report that contains reproducible R commands in both the Rmd file and in the html document [4]. This enables complete reproducibility of statistical results and only requires packaging the R markdown file along with the data.
Detailed usage instructions are coming soon.
Meanwhile, follow the bstr tutorial slides for common use cases and for instructions on how to run the software.
References
- Shattuck DW et al. (2002) BrainSuite: An Automated Cortical Surface Identication Tool Medical Image Analysis, 8(2):129-142.
- Joshi AA et al. (2007) Surface-Constrained Volumetric Brain Registration Using Harmonic Mappings IEEE Trans. on Medical Imaging 26(12):1657-1669.
- Bhushan C et al. (2015) Co-registration and distortion correction of diffusion and anatomical images based on inverse contrast normalization. Neuroimage (7):115:269-80.
- Xie Y (2017) Dynamic Documents with R and knitr. Chapman and Hall/CRC.