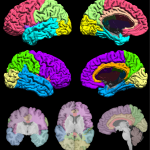
Surface-constrained Volume Registration (SVReg)
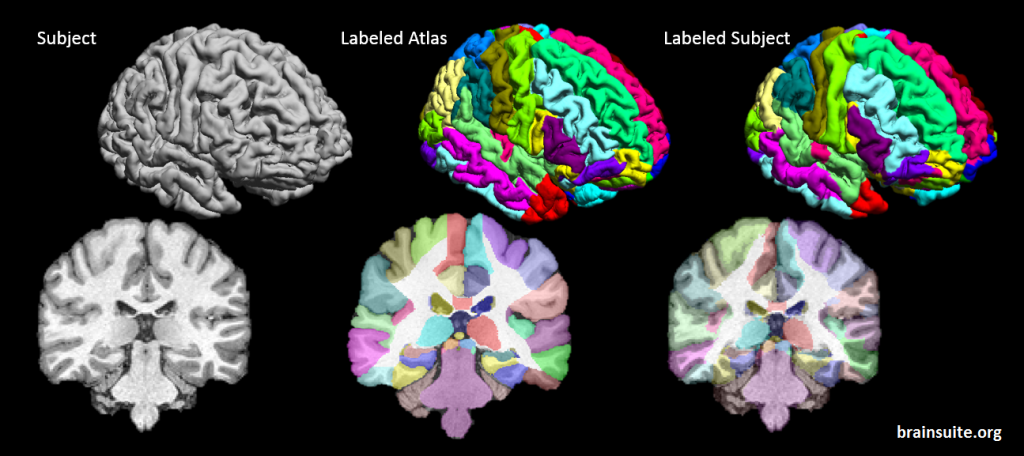
SVReg stands for Surface-constrained Volume Registration. SVReg is an algorithm and a software tool for co-registration of human-brain MR images. It uses anatomical information from both the surface and volume of the brain images for accurate automated co-registration which allows consistent surface and volume mapping to a labeled atlas. Brain labeling is a necessary preprocessing step for several studies looking at group comparisons and regional brain analysis.
Registration Model
SVReg utilizes a series of multi-step registration and refinement processes based on several consideration of useful morphological features and known variation of human brain anatomy. It has the ability to analyze both normal and abnormal brains of subjects starting at least 4 years old, even working well with lesion brains.
Deformation map from atlas to subject
By default SVReg will output:
- Labeled brain surfaces and volumes into segmentations of ~90 ROIs
- Computation of GM, WM, CSF, total volume, average cortical thickness, and surface area of each ROI
- Vertex-wise cortical thickness mapped to atlas surfaces
- Delineation of up to 76 sulci on the mid-cortical surface
Optional Customization
Users may choose to run SVReg through the GUI or through command-line for batch processing and customization.
Users have the option to choose from a number of computational flags or designate manual inputs for highly controlled registration including manual masks, sulcal inputs, and atlases. SVReg can additionally be run in a one step process or be run piecemeal by executing individual modules.
Additional Tools
Additional tools are provided for users who would like to further analyze SVReg.
Our data processing package includes stand-alone functions to read and write SVReg outputs and main scripts to perform the following functions:
- ROI-wise group comparison
- Vertex-wise group comparison
- Surface smoothing
- Regenerating statistics after manual corrections
- Creating custom atlas
- Transforming data to atlas space
- Edit ROIs on surfaces and transfer edits to volume labels
Additionally we provide tools for:
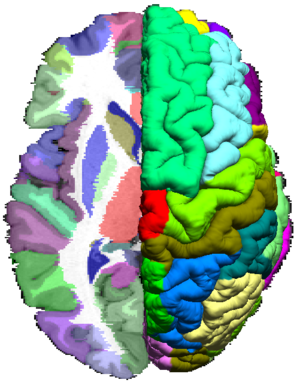

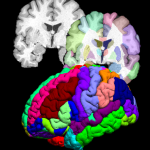
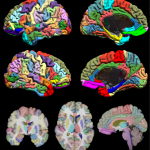
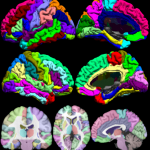 The BrainSuiteAtlas1 is based on the Colin27 atlas and is a stereotaxic average of 27 scans of an individual which is widely used by the neuroscience community.
The BrainSuiteAtlas1 is based on the Colin27 atlas and is a stereotaxic average of 27 scans of an individual which is widely used by the neuroscience community.