USCBrain Atlas
A Hybrid High-Resolution Anatomical MRI Atlas with Subparcellation of Cortical Gyri using Resting fMRI
Developed by Anand A. Joshi, Soyoung Choi, Jessica L. Wisnowski, Justin P. Haldar, David W. Shattuck, Hanna Damasio, and Richard M. Leahy
updated 12 September 2020

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License
When using the USCBrain Atlas in your published work, please cite the following reference paper:
Atlas Description
The USCBrain atlas is a new high-resolution single-subject atlas, constructed using both anatomical and functional information to guide the parcellation of the cortex.
The USCBrain atlas consists of consistent volumetric and surface-based parcellations and labels. The volumetric component of the atlas contains a total of 159 regions of interest (130 cortical, 29 noncortical) defined with respect to the volumetric space. The surface-based component of the atlas as the same 130 sub-gyral (cortical) regions delineated and labeled in addition to the identification of 76 sulcal curves and delineation of gyral crowns and sulcal banks for each gyrus.
We also provide uncertainty maps that give probabilistic parcellations. The USCBrain atlas was developed to be used with BrainSuite to register individual structural T1-weighted scans to this atlas and perform structural labeling. The atlas can also be used with other brain imaging software packages, including FSL and FreeSurfer. Instructions for using the atlas with BrainSuite, FreeSurfer, and FSL are available here.
Anatomical Parcellation
A high-resolution 3D MPRAGE scan (TE=4.33 ms; TR=2070 ms; TI=1100ms; flip angle=12 degrees; resolution=0.547×0.547×0.802 mm) was acquired on a 3T Siemens MAGNETOM Tim Trio using a 32-channel head coil. Data were acquired 5 times and averaged. The subject is a typical right-handed woman in her mid-thirties with a brachiocephalic brain and no rare anomalies.
Preprocessing and labeling of the MPRAGE image was performed using BrainSuite [1,2]. This MRI volume was used to generate the BCI-DNI atlas as described here.
Functional Subparcellation
Functional subparcellations of the gyral ROIs were then generated from 40 minimally preprocessed rfMRI datasets from the Human connectome database [3]. Gyral ROIs were transferred from the BCI-DNI atlas to the 40 subjects using the HCP gray-ordinate space as a reference. For each subject, each gyral ROI was subdivided using the rfMRI data by applying the normalized cuts spectral clustering algorithm to a similarity matrix computed from the correlation between voxels [4]. The number of subdivisions was chosen based on a Silhouette analysis [5]. The subparcellations were then transferred back to the original cortical mesh to create the USCBrain atlas with a total of 65 cortical regions per hemisphere. Probabilistic map of the subdivisions was generated by measuring agreement of the subdivisions across the 40 subjects [6]. The probability maps are stored in .mod files.
Usage
Detailed instructions for using the USCBrain atlas with BrainSuite, FreeSurfer, and FSL are available here.
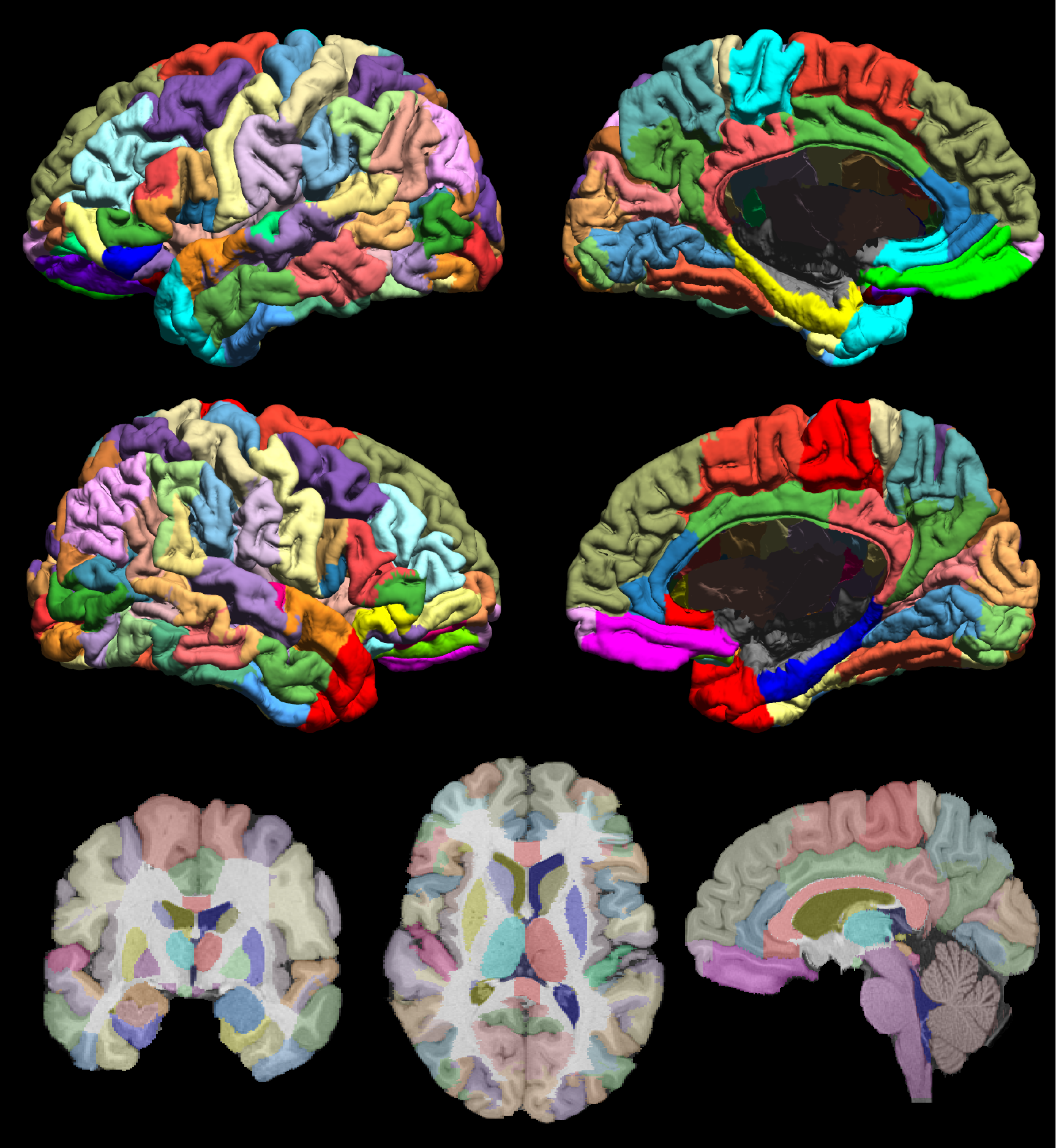
Fig. 1 The USCBrain atlas with 130 cortical regions of interests subdivided using rfMRI data. The figure shows the mid-cortical surface of the atlas, color coded for each sub-parcellated region. Labeled left (top row) and right (bottom row) hemisphere and mesial (right column) and lateral (left column) mid-cortical surfaces. (bottom row) Single-slice skull-stripped MPRAGE image with labels overlaid on coronal (left), axial (middle) and sagittal (right) orientation.
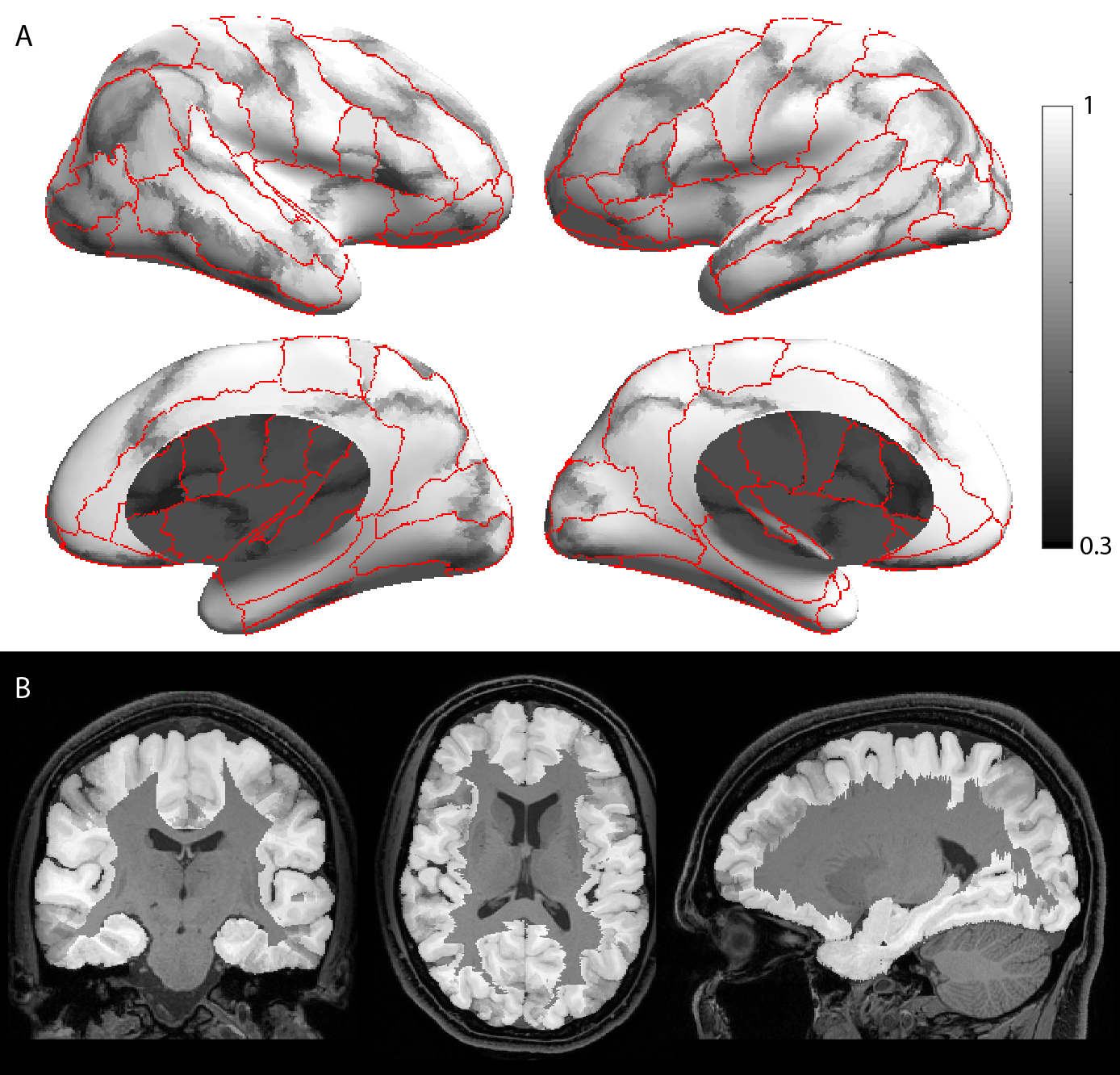
Fig. 2. Grayscale modulated plot showing agreement of the USCBrain atlas labels across the 40 subjects. White (value of 1) indicates perfect agreement at that vertex. The probability maps were generated on the surface as shown in (a) and transferred to the volume as shown in (b). Red curves indicate ROI boundaries of the BCI-DNI atlas.
References
1. Damasio, H., (1995), ‘Human brain anatomy in computerized images’, Oxford university press.
2. Joshi, A., (2012), ‘A method for automated cortical surface registration and labeling’, International Workshop on Biomedical Image Registration, pp 180-189.
3. Glasser, M. (2013), ‘The minimal preprocessing pipelines for the human connectome project’, NeuroImage, vol. 80, pp. 105–124
4. Shi, J. (2000), ‘Normalized cuts and image segmentation’, IEEE Trans on PAMI, 22(8), 888-905.
5. Rousseeuw P. (1987), ‘Silhouettes: a Graphical Aid to the Interpretation and Validation of Cluster Analysis’, Computational and Applied Mathematics 20: 53-65.
6. Hubert, L., (1985), ‘Comparing partitions’, Journal of Classification, Vol 2, pp 193.
7. Joshi, A et al. (2017), ‘A Whole Brain Atlas with Sub-parcellation of Cortical Gyri using Resting fMRI’, SPIE Medical Imaging, 101330O-101330O-9.
8. Joshi, A. ‘USCBrain Atlas: A Volumetric and Surface Atlas Delineated by Anatomical and Functional MRI’, OHBM 2017. e-poster