Overview
Surface-volume registration (SVReg) is an analysis sequence in part of the BrainSuite MR image analysis software.
This documentation will briefly describe the executables and files that are part of SVREG software. SVReg can be performed by using GUI interface. However, if you would like to become an advanced user for more specialized performance, this documentation can be helpful in both understanding how the software performs, how to deconstruct and utilize its separate components, set user specific parameters, and use optional flags.
SVREG consists of a series of modules that can either be called by using the main script through the command line (summarized in the flowchart shown right of the page)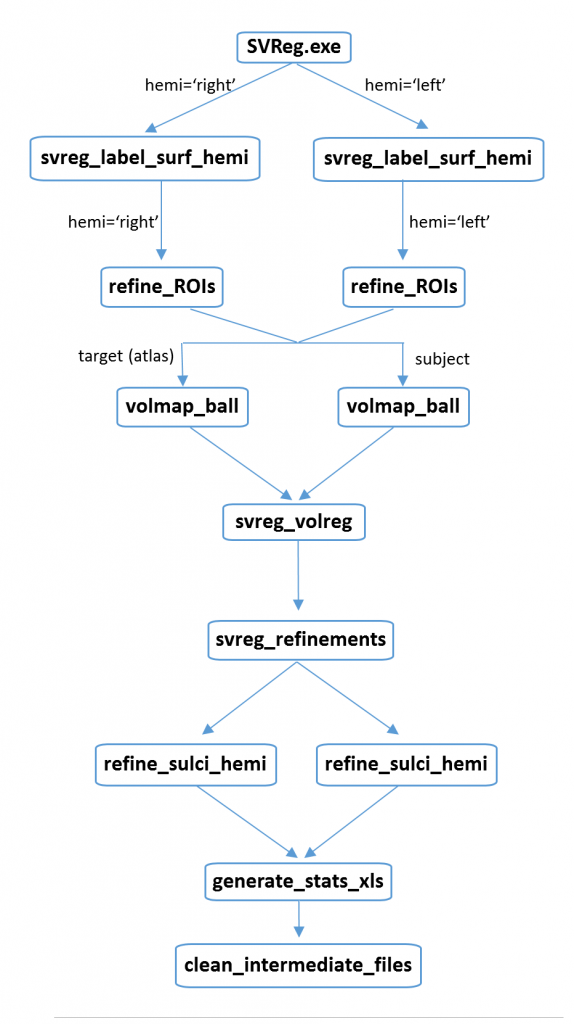 , or through the BrainSuite GUI interface. This document first describes the main script and then describes the separate modules individually.
, or through the BrainSuite GUI interface. This document first describes the main script and then describes the separate modules individually.
Before you begin
- System Requirements
- SVReg requires write permission to the working directory. Make sure to call your data from the appropriate directory.
- SVReg reads, writes, and deletes data quickly. It is therefore recommended that your working directory is not in a syncing environment (such as dropbox). It is also not recommended to work from an external harddrive that may easily be disconnected unless you have a secure, high-speed peripheral connection.
- SVReg requires ~4GB of physical memory to run successfully. SVReg will halt if memory runs out.
- SVReg DirectoryYour SVReg directory will be found in your BrainSuite directory indicated during installation. For most people this directory will be “C:\Program Files\BrainSuite16a1\svreg” where you’ll find a few subdirectories.
- Executable modules will be found in svreg’s subdirectory “bin”
“C:\Program Files\BrainSuite16a1\svreg\bin” - Two single subject atlas templates are provided:
- BrainSuiteAtlas1
- BCI-DNI_brain_atlas
- Executable modules will be found in svreg’s subdirectory “bin”
- SVReg Nomenclature
- We will assume that the full path to your data is located in directory:
C:\Users\BrainSuite_User\Documents\sample_mri - Your unique data identifier will be called your fileprefix. For your original T1 nifti file <Subject1.nii.gz>, your fileprefix will be:
Subject1 - The subbasename will then be:
C:\Users\BrainSuite_User\Documents\sample_mri\Subject1
Only files with the indicated subbasename will be called during SVReg processing unless noted otherwise by appropriate flags. An error message will be shown if required file inputs are not found.
- The atlasbasename will refer to one of the two atlases provided (unless the users decides to use a custom atlas):
- C:\Program Files\BrainSuite16a1\svreg\BrainSuiteAtlas1\mri
- C:\Program Files\BrainSuite16a1\svreg\BCI-DNI_brain_atlas\BCI-DNI_brain
- We will assume that the full path to your data is located in directory:
Modules Usage and Descriptions
- svreg: The Main ScriptDescription:
- This is the main standalone script, compiled and includes the modules that are documented below. Note that the module executables are not necessary for the execution of the svreg, and all the modules are included in the main executable. This is the script to be used when user wants to run the full SVReg sequence using one command. All the modules will run sequentially. The flags are described later in the document. The program will check if the necessary files are present and if not the missing files will be indicated in an error message.
Usage:
- svreg.exe subbasename atlasbasename [-flag1 -flag2 …]
Example Usage:
- svreg.exe C:\Users\BrainSuite_User\Documents\sample_mri\Subject1 C:\Program Files\BrainSuite16a1\svreg\BrainSuiteAtlas1\mri –v2
Input files required:
- subbasename.bfc.nii.gz
- subbasename.cerebrum.mask.nii.gz
- subbasename.cortex.dewisp.mask.nii.gz
- subbasename.hemi.label.nii.gz
- subbasename.mask.nii.gz
- subbasename.pvc.frac.nii.gz
- subbasename.left.inner.cortex.dfs
- subbasename.left.pial.cortex.dfs
- subbasename.pial.cortex.dfs
- subbasename.right.inner.cortex.dfs
- subbasename.right.pial.cortex.dfs
- subbasename.warp
Outputs
- Files are written out with the same input subbasename.
- Descriptions are detailed throughout the document and a summary of output files can be found
- .
- svreg_label_surf_hemiDescription:
- This is the main routine for surface registration. It performs flattening of the surfaces, L2 registration and curvature registration. First the surface is smoothed to create a multiresolution representation of the surface. The 3D coordinates of the smoothed surface are aligned to perform the initial registration. The L2 based registration is followed by p-harmonic mapping to a square. This is then followed by computation of curvatures for the multiresolution surfaces and a curvature based registration.
Results:
- Initial registration of the subject to atlas surfaces, labels/segmentation, and sulcal curves.
Usage:
- svreg_label_surf_hemi.exe subbasename atlasbasename hemisphere (‘left’ or ‘right’) [flag1 flag2 …]
Example Usage:
- svreg_label_surf_hemi.exe C:\Users\BrainSuite_User\Documents\sample_mri\Subject1 C:\ProgramFiles\BrainSuite\SVREG\BrainSuiteAtlas1\mri left -r
Input files required:
- subbasename.hemi.inner.cortex.dfs
- subbasename.hemi.pial.cortex.dfs
- subbasename.tmp.svreg/subbasename.target.hemi.inner.cortex.dfs
- subbasename.tmp.svreg/subbasename.target.hemi.pial.cortex.dfs
- subbasename.warp
Flags:
- Sulcal constraints:
It is possible to use sulcal constraints for performing the cortical registration using the –cur flag.
-cur sulc_name.dfc #sul1 #sul2… This flag is used in svreg_label_surf_hemi script. One can specify the name of the dfc file and the sulcal numbers to be used as constraints. - Message and time stamp flags: -t –v -m
Outputs:
- subbasename.svreg.tmp/subbasename.svreg.version
- subbasename.svreg.tmp/subbasename.hemi.inner.cortex.reg.dfs
- subbasename.svreg.tmp/subbasename.hemi.pial.cortex.reg.dfs
- subbasename.svreg.tmp/subbasename.hemi.mid.cortex.reg.dfs
- subbasename.svreg.tmp/subbasename.target.hemi.mid.cortex.svreg.dfs
- subbasename.svreg.tmp/subbasename.hemi.mapped.all.dfc
- subbasename.svreg.tmp/subbasename.hemi.mapped.dfc
- subbasename.svreg.tmp/subbasename.target.hemi.mapped.all.dfc
- subbasename.svreg.tmp/subbasename.target.hemi.dfc
- subbasename.svreg.tmp/subbasename.target.hemi.all.dfc
- subbasename.svreg.tmp/subbasename.hemi.mapped.dfc
- subbasename.svreg.tmp/subbasename.hemi.mapped.all.dfc
- subbasename.svreg.tmp/subbasename.target.warp
- atlas.hemi.mid.cortex.svreg.dfs
- subbasename.hemi.inner.cortex.init.svreg.dfs
- subbasename.hemi.pial.cortex.init.svreg.dfs
- subbasename.hemi.mid.cortex.init.svreg.dfs
- Sulcal constraints:
- refine_ROIs2Description:
- This function refines the ROI boundaries on the surface using the geodesic curvature flow. The ROI boundaries are modified such that they conform to the underlying anatomical curvature. The surfaces are then recolored according to the labels.
- There are a subset of ROI boundaries that are excluded from these refinement steps. (for example: boundary between the cingulate [184/185] and superior frontal gyrus [120/121] is excluded)
Results:
- Refined labeling of surface labels/segmentations using individual subject’s local surface-based morphological features.
Usage:
- refine_ROIs2.exe subbasename hemi [-flag1 -flag2 …]
Example Usage:
- refine_ROIs2.exe C:\Users\BrainSuite_User\Documents\sample_mri\Subject1 left –r
Flags:
- Message and time stamp flags: -t –v -m
Input files required:
- subbasename.hemi.mid.cortex.init.svreg.dfs
Outputs:
- subbasename.hemi.mid.cortex.svreg.dfs
- volmap_ballDescription:
- Maps the brain surface and volume to the unit ball. This is done for both the hemispheres by using a PCG routine.
Results:
- Unitball mapping of the subject’s brain surfaces and volume.
Usage:
- volmap_ball.exe subbasename atlasbasename 1 [–flag1 -flag2 …]
Example Usage:
- volmap_ball.exe C:\Users\BrainSuite_User\Documents\sample_mri\Subject1 1
Flags:
- Message and time stamp flags: -t –v -m
Input files required:
- subbasename.cortex.dewisp.mask.nii.gz
- subbasename.tmp.svreg/subbasename.target.cortex.dewisp.mask.nii.gz
- subbasename.tmp.svreg/subbasename.left.inner.cortex.reg.dfs
- subbasename.tmp.svreg/subbasename.right.inner.cortex.reg.dfs
- subbasename.tmp.svreg/subbasename.target.left.inner.cortex.reg.dfs
- subbasename.tmp.svreg/subbasename.target.right.inner.cortex.reg.dfs
- subbasename.tmp.svreg/subbasename.left.pial.cortex.reg.dfs
- subbasename.tmp.svreg/subbasename.right.pial.cortex.reg.dfs
- subbasename.tmp.svreg/subbasename.target.left.pial.cortex.reg.dfs
- subbasename.tmp.svreg/subbasename.target.right.pial.cortex.reg.dfs
Outputs:
- subbasename.tmp.svreg/subbasename_unitball_map.mat,
- subbasename.tmp.svreg/subbasename.target_unitball_map.mat
- svreg_volregDescription:
- Performs inversion of the p-harmonic spherical maps followed by initial mapping from subject brain to the atlas brain.
- A deformation field is calculated using the surface registration results from the previous step. Affine deformation is found by fitting an affine registration matrix between the pial cortex surface coordinates of the subject and atlas. The field 15 voxels outside of the brain mask image (mask.nii.gz) is used.
- A second deformation field is calculated between the atlas and subject using the boundaries of the white matter mask (cortex.dewisp.mask.nii.gz) using harmonic mapping to the unit ball. The hippocampus and structures inferior to this region are excluded from this mapping process. The map within the boundaries of the white matter is used and then extends out to the boundary of the pial cortex.
- Two separate deformation fields are calculated by using the constraints of the white matter and the pial surface between the atlas and subject then combined in 3D using a cost function that utilizes a Laplacian operator.
This is then followed by an intensity registration step using pvc.frac that refines the deformation map based on volumetric morphological features.
An intensity-based registration is performed to refine registration of non-cortical structures, including the subcotex, extra-cerebral structures, and hippocampus. This region is constrained by the boundary of the WM with addition of the brainstem and cerebellum dilated by 3 voxels.Results:
- Refined volumetric deformation field between the subject and atlas.
Usage:
- svreg_volreg.exe subbasename atlasbasename [–flag1 –flag2 …]
Example Usage:
- svreg_volreg.exe C:\Users\BrainSuite_User\Documents\sample_mri\Subject1 -tv2
Flags:
- Message and time stamp flags: -t –v -m
Input files required:
- subbasename.cortex.dewisp.mask.nii.gz
- subbasename.tmp.svreg/subbasename.target.cortex.dewisp.mask.nii.gz
- subbasename.cortex.dewisp.mask.nii.gz
- subbasename.tmp.svreg/subbasename.target.cortex.dewisp.mask.nii.gz
- subbasename.tmp.svreg/subbasename.left.inner.cortex.reg.dfs
- subbasename.tmp.svreg/subbasename.right.inner.cortex.reg.dfs
- subbasename.tmp.svreg/subbasename.target.left.inner.cortex.reg.dfs
- subbasename.tmp.svreg/subbasename.target.right.inner.cortex.reg.dfs
- subbasename.tmp.svreg/subbasename.left.pial.cortex.reg.dfs
- subbasename.tmp.svreg/subbasename.right.pial.cortex.reg.dfs
- subbasename.tmp.svreg/subbasename.target.left.pial.cortex.reg.dfs
- subbasename.tmp.svreg/subbasename.target.right.pial.cortex.reg.dfs
- subbasename.tmp.svreg/subbasename_unitball_map.mat,
- subbasename.tmp.svreg/subbasename.target_unitball_map.mat
- subbasename.hemi.label.nii.gz
- subbasename.tmp.svreg/subbasename.target.hemi.label.nii.gz
Outputs:
- subbasename.tmp.svreg/subbasename.surfreg.label.nii.gz
- subbasename.tmp.svreg/subbasename.surfreg.map.nii.gz
- subbasename.tmp.svreg/subbasename.surfreg.nii.gz
- subbasename.svreg.map.nii.gz
- subbasename.inv.svreg.map.nii.gz
- subbasename.tmp.svreg/subbasename.svreg.nii.gz
- subbasename.tmp.svreg/subbasename.inv.svreg.nii.gz
- svreg_refinementsDescription:
- Refines the maps and volumetric labels based on surface based refinement labels outputted from module refine_ROIs2 (subbasename.hemi.svreg.dfs). Additionally checks and corrects the topology of the volumetric and surface based ROIs.
- Topology of the labels are checked (subbasename.tmp.svreg/subbasename.svreg.ref.label.nii.gz) and corrected if wrong. (Checks for isolated voxels and disconnected labels, holes inside labels)
- The inner boundary of the cortical labels cutting into deep white matter is smoothed by median filtering (subbasename.svreg.label.nii.gz). incorrect
Results:
- Refined deformation field of atlas to subject, volumetric labels/segmentation, and labeled/segmented surfaces of subject.
Usage:
- svreg_refinements.exe subbasename atlasbasename [-flag1 -flag2 …]
Example Usage:
- svreg_refinements.exe C:\Users\BrainSuite_User\Documents\sample_mri\Subject1 C:\Users\BrainSuite_User\Documents\sample_mri\Subject1.target -r
Flags:
Input files required:
- subbasename.hemi.mid.cortex.svreg.dfs
- subbasename.hemi.inner.cortex.svreg.dfs
- subbasename.hemi.pial.cortex.svreg.dfs
- subbasename.svreg.tmp/subbasename.svreg.label.nii.gz
- subbasename.svreg.map.nii.gz
Outputs:
- subbasename.svreg.tmp/subbasename.svreg.wm.label.nii.gz
- subbasename.svreg.tmp/subbasename.svreg.label.nii.gz
- subbasename.svreg.tmp/subbasename.jacobian.nii.gz
- subbasename.svreg.tmp/subbasename.svreg.corr.label.nii.gz)
- subbasename.hemi.mid.cortex.svreg.dfs
- subbasename.hemi.inner.cortex.svreg.dfs
- subbasename.hemi.pial.cortex.svreg.dfs
- refine_sulci_hemiDescription:
- Refines the sulcal curves by using geodesic curvature flow so that they conform to the underlying geometry of the mid-cortical surface.
Results:
- Refined sulcal curves
Usage:
- refine_sulci_hemi.exe subbasename hemi [–flag1 -flag2 …]
Example Usage:
- refine_sulci_hemi.exe C:\Users\BrainSuite_User\Documents\sample_mri\Subject1 left –gui
Flags:
Input files required:
- subbasename.svreg.tmp/subbasename.hemi.mapped.dfc
- subbasename.svreg.tmp/subbasename.hemi.mid.cortex.reg.dfs
Outputs:
- subbasename.hemi.mapped.refined.dfc
- generate_stats_xlsDescription:
- Generates delimited .txt file representing ROIwise average gray matter thickness and gray matter volumes for the cortical ROIs and volumes of the subcortical ROIs. This generates a formatted text file which can be imported into excel if required.
Results:
- Statistics.
Usage:
- generate_stats_xls.exe subbasename
Example Usage:
- generate_stats_xls.exe C:\Users\BrainSuite_User\Documents\sample_mri\Subject1
Flags:
Input files required:- subbasename.svreg.tmp/subbasename.left.mid.cortex.reg.dfs
- subbasename.svreg.tmp/subbasename.right.mid.cortex.reg.dfs
- subbasename.svreg.label.nii.gz
- subbasename.pvc.frac.nii.gz
Outputs:
- subbasename.roiwise.stats.txt
- clean_intermediate_filesDescription:
- This function deletes intermediate files that were copied for the svreg sequence. If –k flag was used, it keeps all the intermediate files which could be useful for debugging.
- This module should be used after executing all other modules.
Results:
- Deletes intermediate files which are usually unnecessary for further analysis.
Usage:
- clean_intermediate_files.exe subbasename
Example Usage:
- clean_intermediate_files.exe C:\Users\BrainSuite_User\Documents\sample_mri\Subject1
Temporary folder subbasename.svreg.tmp is deleted:
- thicknessPVCDescription:
- This script represents our new code for computation of cortical thickness using partial tissue fraction volume. It also stores this thickness computed into separate hemisphere files and subject thickness mapped to the atlas hemisphere surfaces. This script needs to be run from the command line separately and is not run through the main SVReg sequence.
- This module should be used after executing the BrainSuite and SVReg sequence.
Results:
- Computes cortical thickness using partial tissue fractions. This thickness measure is then transferred to the atlas surface to facilitate population studies.
Usage:
- thicknessPVC.exe subbasename
Example Usage:
- thicknessPVC.exe C:\Users\BrainSuite_User\Documents\sample_mri\Subject1
Input files required:
- subbasename.inner.cortex.dfs
- subbasename.pial.cortex.dfs
- subbasename.cortex.dewisp.mask.nii.gz
- subbasename.cerebrum.mask.nii.gz
- subbasename.pvc.frac.nii.gz
- atlas.left.mid.cortex.svreg.dfs
- atlas.right.mid.cortex.svreg.dfs
Output files:
- subbasename.GM_frac.nii.gz
- subbasename.heat_map_sol.nii.gz
- subbasename.pvc-thickness_0-6mm.isosurface.dfs
- subbasename.pvc-thickness_0-6mm.mid.cortex.dfs
- subbasename.heat_sol_comp.mat
- subbasename.pvc-thickness_0-6mm.left.mid.cortex.dfs
- subbasename.pvc-thickness_0-6mm.right.mid.cortex.dfs
- atlas.pvc-thickness_0-6mm.left.mid.cortex.dfs
- atlas.pvc-thickness_0-6mm.right.mid.cortex.dfs
:
- svreg_smooth_surf_functionDescription:
- Performs smoothing of a surface based function.
- This function is used to smooth function defined on surface, such as cortical thickness or curvature. Surface based smoothing is typically required for compensating for the results of mis-registration.
Results:
- The output dfs file contains smoothed function and also the surface on which the function was smoothed.
Usage:
- svreg_smooth_surf_function.exe in_surf func_surf out_surf param(optional)
Example Usage:
- svreg_smooth_surf_function.exe Subject1_mid_left.dfs Subject1_mid_left_function.dfs Subject1_mid_left_function_out.dfs param
Input files required:
- input_surf.dfs: This is the surface on which smoothing will be performed.
- input_surf_function.dfs: .attributes field of this file contains the function on the input surface. The input_surf.dfs and input_surf_function.dfs could be same.
- output_surf.dfs: This is the output surface file that contains smoothed function in the attributes field.
- param (optional): This parameter contains smoothing parameter roughly, kernel std dev in mm. Default value is 10
- svreg_make_atlasDescription:
- This function prepares a new atlas from a given subject.
- Users can create their own atlas and then use with SVReg.
To create a new atlas, function takes two arguments: subbasename and atlasbasename. subbasename is the new atlas that we want to create. We assume that the subject specified in subbasename is processed using brainsuite and svreg. atlasbasename is the atlas from BrainSuite that was used to process this subject. This is required in order for the script to copy a few files (.xml,.mat) into the new subject’s directory. If you want to perform manual editing of the labels, then please edit subbasename.svreg.label.nii.gz before running the script. The new atlas folder should be copied in the SVReg directory and kept with other atlas folders.Results:
- The subject directory is updated with all the files and subbasename now can be used as a new atlasbasename in svreg execution.
Usage:
- svreg_make_atlas.exe subbasename atlasbasename flag
Input Files Required:
subbasename: File prefix of the subject that you want to convert into atlas.
atlasbasename: atlas from BrainSuite that was used to process this subject.
flag: -E means volumetric labels are manually edited.Also see here.
- svreg_apply_mapDescription:
- This script applies a map generated by svreg to a given volume.
- SVReg generates svreg.map and svreg.inv.map. These two maps respectively allow users to warp data from atlas to subject space, and subject space to atlas space respectively. This data can be scalar valued data such as T1 image or FA image, or it can be a tensor data such as DTI data. In case of DTI data, a the tensors are reorieneted using PPD (preservation of principle direction) algorithm.
Results:
- The warped image is stored with a given file name
Usage:
- svreg_apply map.exe map data output target
Input Files Required:
map: This file contains a svreg map. (e.g. subbasename.svreg.map.nii.gz).
data: File containing the data to be warped
output: name of the output file
target: This file is used for getting dimensions and pixel resolution of the target image space.
- generate_vol_param_stats_xlsDescription:
- Calculate volumetric and surface stats (svreg). This program calculates the mean thickness, grey matter volume, white matter volume, CSF volume, total volume, and the cortical area of SVReg’s labeled regions of interest. It then saves the results in a csv file.
Results:
- Stats in a txt file
Usage:
- generate_vol_param_stats_xls.sh subject fileprefix param_nii_file [flags]
Input Files Required:
subject_fileprefix path and filename prefix of the subject’s output
from BrainSuite’s Cortical Surface Extraction
Sequence
param_nii_file File that contains scalar map
optional flags:
-v# Controls the verbosity of output messages (# is 0, 1, or 2)
-r Use if svreg sequence was run with refinement
- svreg_resampleDescription:
- This function resamples 3D or 4D input NIFTI-1 file.
- This function can be used to resample a given NIFTI-1 file to a specified resolution or size. The function can resample both 3D and 4D files.
Results:
- The resampled file in NIFTI-1 format
Usage:
- svreg_resample.exe in_file out_file [dim] [dx] [dy] [dz] [method] [extrapval]
Inputs:
in_file: input nifti file in nii.gz file format
out_file: output nifti file in nii.gz format
[dim]: if this is ‘-size’ output size is specified
if this is ‘-res’ output pixel resolution is specified
default is -res 1 1 1 (istotropic sampling)
[dx],[dy],[dz]: if the dim=’-size’, then this is interpreted as output size
if the dim=-res’, then this is interpred as voxel resolution in x,y,z directions
[method]: interpolation methods: ‘linear’, ‘nearest’,’cubic’,’spline’ the default is ‘linear’.
[extrapval]: Values for extrapolation outside the grid on the boundary. - svreg_smooth_vol_functionDescription:
- This script performs 3d vol smoothing.
- This script performs 3d volumetric smoothing with Gaussian.
Results:
- Smoothed volume
Usage:
- svreg_smooth_vol_function.exe in_file stdx stdy stdz out_file
Inputs:
in_file: input vol file
stdx,stdy,stdz: std dev (in mm) in 3 directions
out_vol: output vol file - svreg_get_mni_talDescription:
- Convert subject’s voxel coordinates to MNI and Talairach coordinates.
- The voxel coordinates can be obtained using the BrainSuite interface. The output are MNI and Talairach coordinates.
Results:
- MNI and Talairach coordinates
Usage:
- svreg_get_mni_tal.exe [subject fileprefix] [atlas fileprefix] [sx] [sy] [sz]
Inputs:
subbasename: subjects base name
atlasbasename: atlas base name
sx,sy,sz: xyz voxel coordinates - svreg_labelwith_atlasDescription:
- This function creates labels using a different atlas.
- This function creates labels using a different atlas. This function works
only for BCI-DNI, USCBrain and USCLobes atlases. Since the underlying
anatomy is the same, you can process data using one brain and use it with
another one.Results:
- MNI and Talairach coordinates
Usage:
- svreg_labelwith_atlas.exe subbasename atlasbasename postfix
Inputs:
subbasename: subjects base name
atlasbasename: atlas base name
postfix: postfix to be appended to the output label files of the subject - svreg_sulcal_mapDescription:
- This function creates sulcal map on the cortex.
- This function works
only for BCI-DNI, USCBrain and USCLobes atlases.Results:
- MNI and Talairach coordinates
Usage:
- svreg_labelwith_atlas.exe subbasename atlasbasename XMLFile XMLSFile threshold
Inputs:
subbasename: subjects base name
atlasbasename: atlas base name
XMLFile: XML file that contains atlas description
XMLSFile: XML file that contains description of sulcal anatomy
threshold: Threshold